Probe CUST_16675_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16675_PI426222305 | JHI_St_60k_v1 | DMT400069500 | TCCTTACTTTCCCTTTCTTGTGTGTAGTGTGATGGTTTTTATCAGAAAATGATATTCCCT |
All Microarray Probes Designed to Gene DMG400027019
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16675_PI426222305 | JHI_St_60k_v1 | DMT400069500 | TCCTTACTTTCCCTTTCTTGTGTGTAGTGTGATGGTTTTTATCAGAAAATGATATTCCCT |
Microarray Signals from CUST_16675_PI426222305
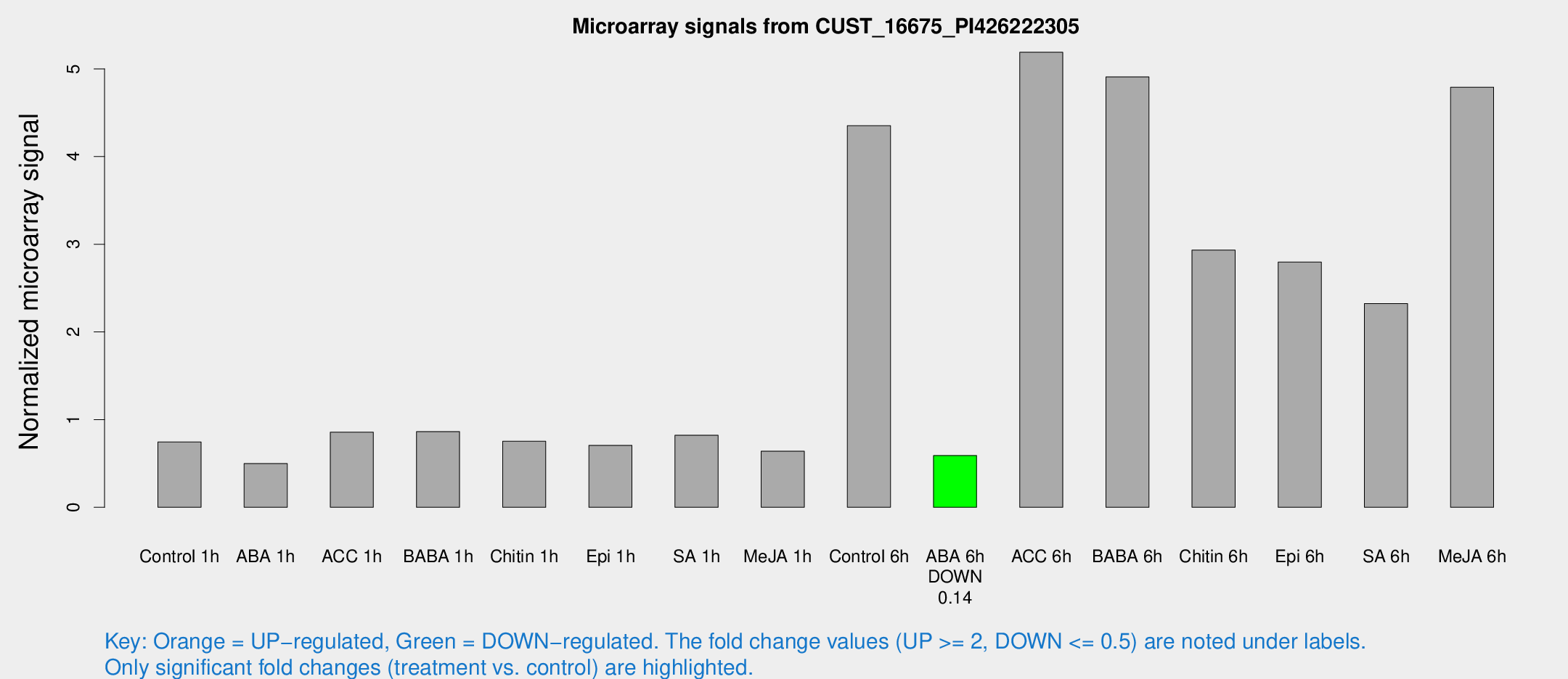
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 958.363 | 80.7106 | 0.745022 | 0.0430919 |
| ABA 1h | 574.422 | 76.0483 | 0.498578 | 0.108266 |
| ACC 1h | 1260.95 | 375.487 | 0.856831 | 0.26107 |
| BABA 1h | 1126.91 | 243.7 | 0.863258 | 0.129082 |
| Chitin 1h | 889.983 | 139.998 | 0.752929 | 0.0888365 |
| Epi 1h | 802.549 | 127.234 | 0.706834 | 0.114712 |
| SA 1h | 1084.26 | 74.5216 | 0.822359 | 0.0755868 |
| Me-JA 1h | 838.113 | 398.031 | 0.640524 | 0.338707 |
| Control 6h | 6402.18 | 1971.27 | 4.35167 | 1.45173 |
| ABA 6h | 808.899 | 76.7353 | 0.590185 | 0.0901298 |
| ACC 6h | 8401.54 | 2275.85 | 5.18981 | 1.45075 |
| BABA 6h | 7053.95 | 487.227 | 4.90765 | 0.473532 |
| Chitin 6h | 4112.25 | 698.97 | 2.93411 | 0.427232 |
| Epi 6h | 4031.98 | 233.019 | 2.79642 | 0.2436 |
| SA 6h | 3356.37 | 1023.78 | 2.32398 | 0.68109 |
| Me-JA 6h | 6169.2 | 652.44 | 4.79089 | 0.886431 |
Source Transcript PGSC0003DMT400069500 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G70370.1 | +3 | 9e-37 | 143 | 71/213 (33%) | polygalacturonase 2 | chr1:26513003-26514998 REVERSE LENGTH=626 |
