Probe CUST_15941_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15941_PI426222305 | JHI_St_60k_v1 | DMT400057821 | CCGGCGCCCATTGTATAGTGTTTTCGAACTTAATTTTCAATCTTCCCCTAAAACATTCTT |
All Microarray Probes Designed to Gene DMG400022450
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15941_PI426222305 | JHI_St_60k_v1 | DMT400057821 | CCGGCGCCCATTGTATAGTGTTTTCGAACTTAATTTTCAATCTTCCCCTAAAACATTCTT |
Microarray Signals from CUST_15941_PI426222305
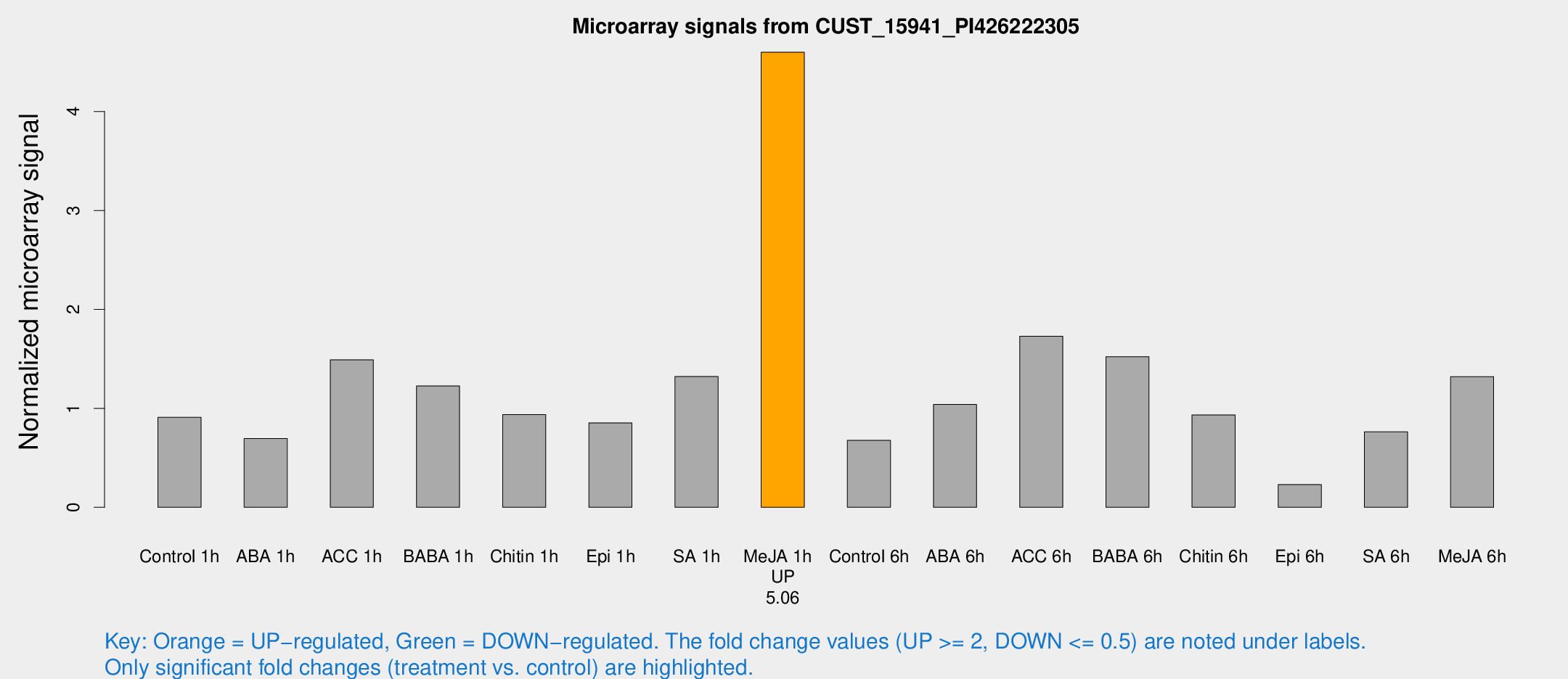
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1129.72 | 287.304 | 0.909492 | 0.183799 |
| ABA 1h | 778.1 | 193.102 | 0.695884 | 0.168061 |
| ACC 1h | 2049.84 | 739.91 | 1.48989 | 0.504706 |
| BABA 1h | 1387.21 | 80.4038 | 1.22675 | 0.15246 |
| Chitin 1h | 1002.32 | 129.643 | 0.937989 | 0.123816 |
| Epi 1h | 887.423 | 147.695 | 0.85341 | 0.118726 |
| SA 1h | 1618.53 | 210.244 | 1.322 | 0.269446 |
| Me-JA 1h | 4427.35 | 404.417 | 4.59919 | 0.265556 |
| Control 6h | 898.186 | 276.069 | 0.676309 | 0.20187 |
| ABA 6h | 1298.57 | 77.8713 | 1.04055 | 0.0601376 |
| ACC 6h | 2356.98 | 300.378 | 1.72836 | 0.147024 |
| BABA 6h | 2231.95 | 777.374 | 1.5211 | 0.508519 |
| Chitin 6h | 1164.04 | 67.4489 | 0.93298 | 0.0539432 |
| Epi 6h | 303.394 | 17.9342 | 0.229755 | 0.0157383 |
| SA 6h | 902.62 | 131.353 | 0.763007 | 0.0487111 |
| Me-JA 6h | 1547.83 | 123.059 | 1.32055 | 0.185976 |
Source Transcript PGSC0003DMT400057821 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
