Probe CUST_15883_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15883_PI426222305 | JHI_St_60k_v1 | DMT400057819 | TTCATCTTCCATATGGCTGAGCGTTTTTTCTTAGGAAAATTAGGCGAGTTCGTAAATATT |
All Microarray Probes Designed to Gene DMG400022448
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15883_PI426222305 | JHI_St_60k_v1 | DMT400057819 | TTCATCTTCCATATGGCTGAGCGTTTTTTCTTAGGAAAATTAGGCGAGTTCGTAAATATT |
Microarray Signals from CUST_15883_PI426222305
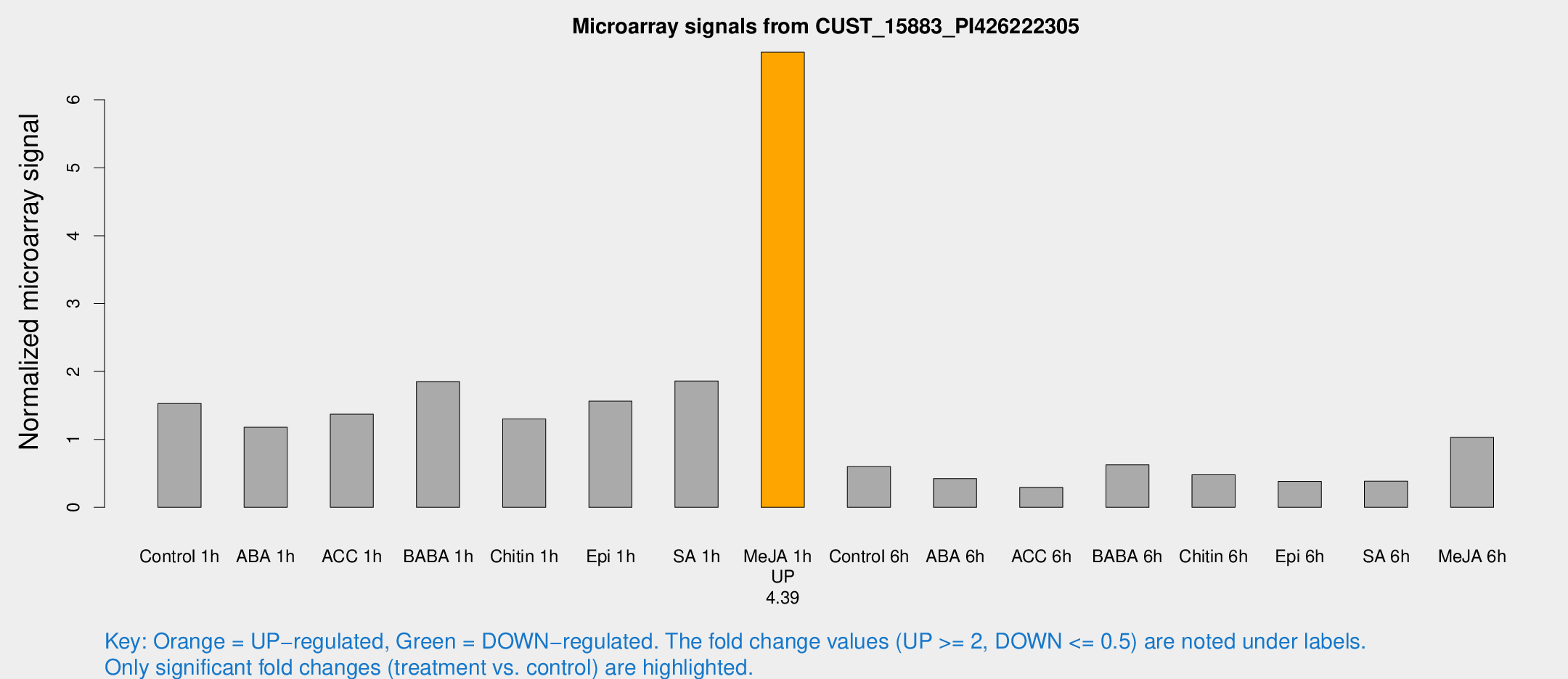
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7099.01 | 1825.42 | 1.52803 | 0.315768 |
| ABA 1h | 5134.46 | 1620.57 | 1.17884 | 0.371623 |
| ACC 1h | 7145.93 | 2619.51 | 1.37166 | 0.500107 |
| BABA 1h | 7991.06 | 1198.83 | 1.85139 | 0.113806 |
| Chitin 1h | 5189.59 | 638.033 | 1.30192 | 0.240123 |
| Epi 1h | 6019.45 | 792.87 | 1.56216 | 0.206405 |
| SA 1h | 8417.57 | 776.875 | 1.85947 | 0.296921 |
| Me-JA 1h | 24109.7 | 2316.57 | 6.70203 | 0.386943 |
| Control 6h | 2728.91 | 532.761 | 0.598089 | 0.0696745 |
| ABA 6h | 1994.56 | 246.156 | 0.423018 | 0.0443398 |
| ACC 6h | 1499.35 | 207.966 | 0.293104 | 0.0581197 |
| BABA 6h | 3276.82 | 845.556 | 0.625142 | 0.169697 |
| Chitin 6h | 2241.63 | 207.963 | 0.477781 | 0.0618302 |
| Epi 6h | 1896.7 | 165.711 | 0.382271 | 0.0504627 |
| SA 6h | 1720.68 | 300.368 | 0.384352 | 0.0299152 |
| Me-JA 6h | 4497.77 | 285.269 | 1.03013 | 0.107624 |
Source Transcript PGSC0003DMT400057819 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
