Probe CUST_15515_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15515_PI426222305 | JHI_St_60k_v1 | DMT400073709 | CAAGAACACACGACTTTGTTCACACATTAGATCCTGTTACTAGCCATTGAATATTATGAT |
All Microarray Probes Designed to Gene DMG400028632
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15515_PI426222305 | JHI_St_60k_v1 | DMT400073709 | CAAGAACACACGACTTTGTTCACACATTAGATCCTGTTACTAGCCATTGAATATTATGAT |
Microarray Signals from CUST_15515_PI426222305
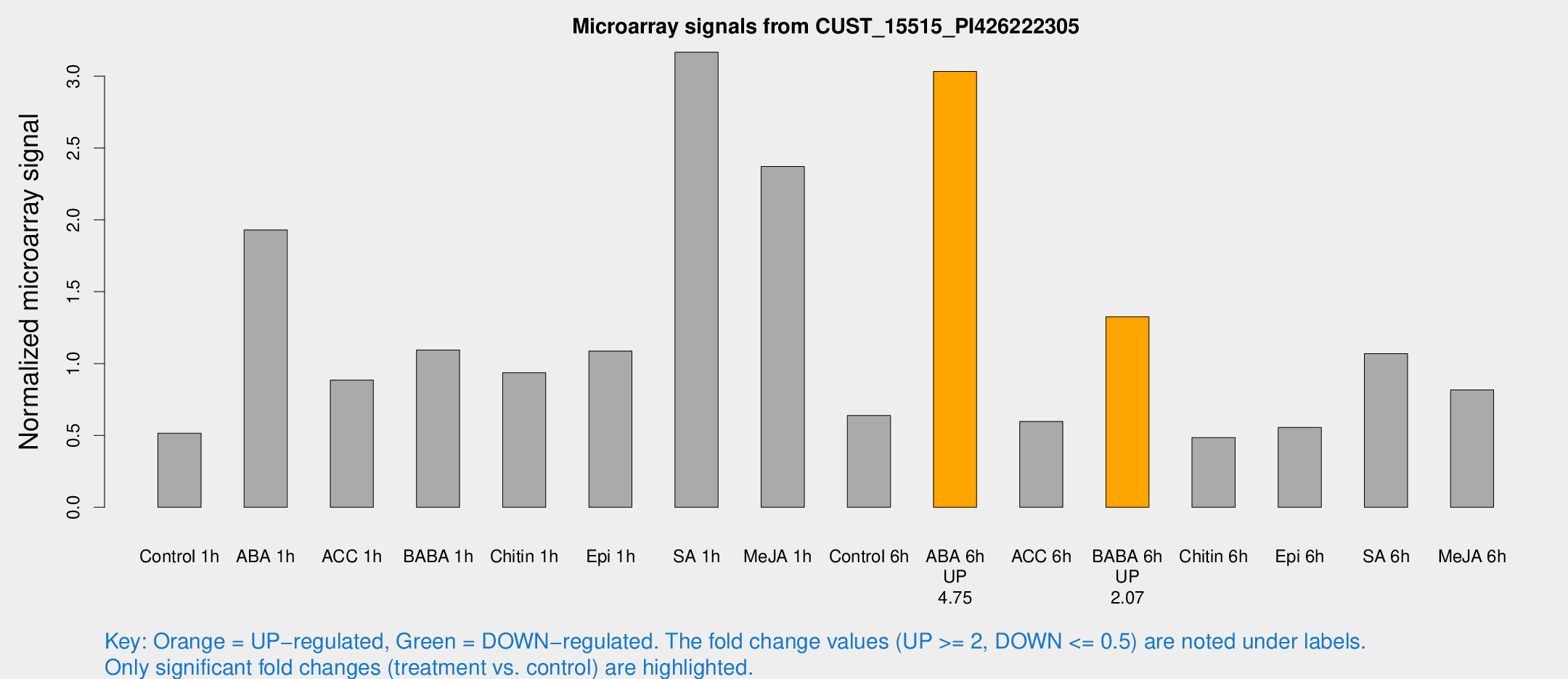
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 522.223 | 62.3566 | 0.514754 | 0.0840885 |
| ABA 1h | 1712.59 | 105.583 | 1.92906 | 0.258306 |
| ACC 1h | 943.187 | 171.159 | 0.884629 | 0.114069 |
| BABA 1h | 1111.82 | 241.479 | 1.09407 | 0.154804 |
| Chitin 1h | 852.446 | 97.6966 | 0.936211 | 0.104646 |
| Epi 1h | 943.819 | 57.3087 | 1.0868 | 0.0940472 |
| SA 1h | 3459.03 | 801.542 | 3.16609 | 0.716796 |
| Me-JA 1h | 2013 | 366.915 | 2.37105 | 0.338242 |
| Control 6h | 666.696 | 133.838 | 0.638734 | 0.0769251 |
| ABA 6h | 3317.34 | 527.87 | 3.03194 | 0.397047 |
| ACC 6h | 696.224 | 91.1168 | 0.596933 | 0.0346594 |
| BABA 6h | 1500.77 | 158.192 | 1.32531 | 0.178488 |
| Chitin 6h | 519.353 | 37.6731 | 0.485472 | 0.0446493 |
| Epi 6h | 642.997 | 98.4682 | 0.555735 | 0.129593 |
| SA 6h | 1278.99 | 509.458 | 1.06965 | 0.414034 |
| Me-JA 6h | 852.063 | 181.437 | 0.816707 | 0.170904 |
Source Transcript PGSC0003DMT400073709 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G36780.1 | +2 | 4e-179 | 520 | 271/489 (55%) | UDP-Glycosyltransferase superfamily protein | chr2:15417618-15419108 REVERSE LENGTH=496 |
