Probe CUST_14540_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14540_PI426222305 | JHI_St_60k_v1 | DMT400066480 | ATCGAAGGCATACACATTTCATTATCGTGTATTGTAAGACTGATTTCCTAGATTTGGTGT |
All Microarray Probes Designed to Gene DMG400025834
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14540_PI426222305 | JHI_St_60k_v1 | DMT400066480 | ATCGAAGGCATACACATTTCATTATCGTGTATTGTAAGACTGATTTCCTAGATTTGGTGT |
Microarray Signals from CUST_14540_PI426222305
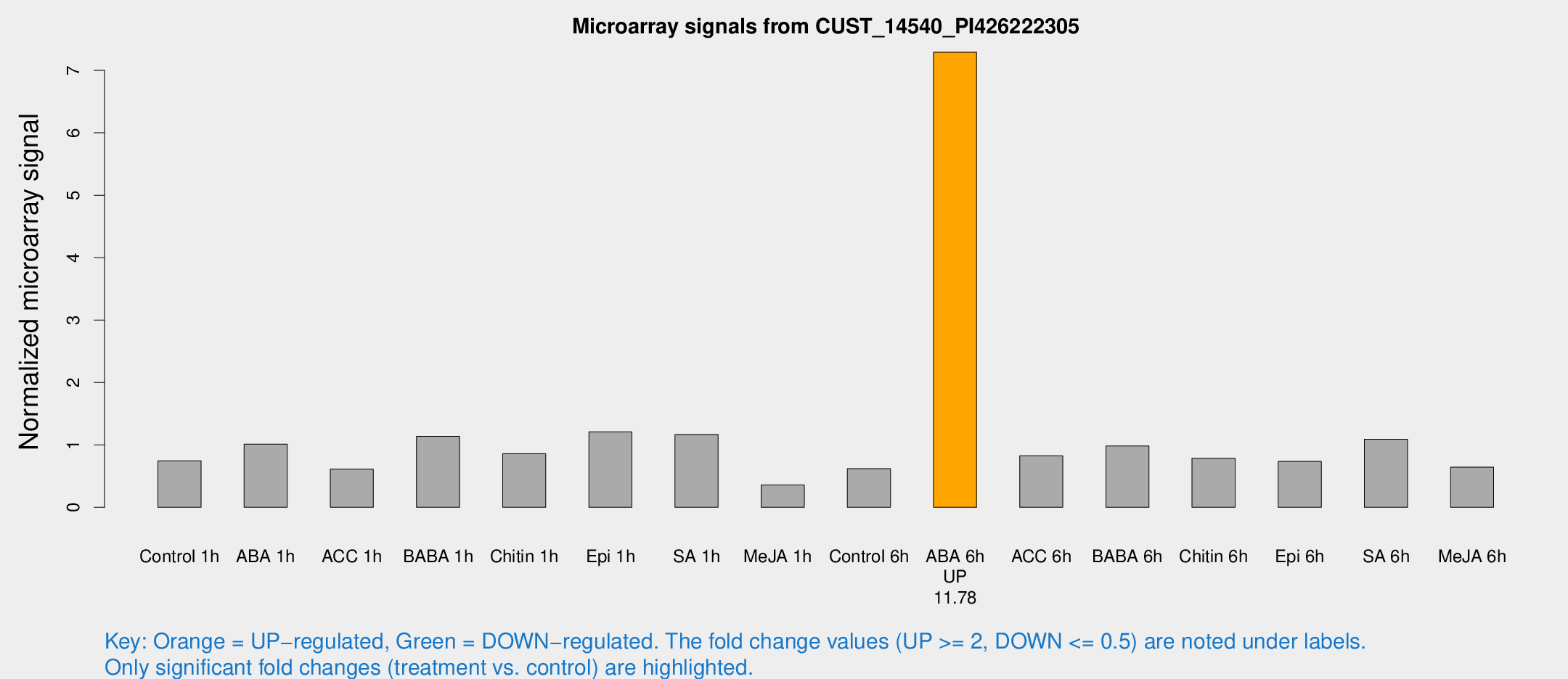
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 20.3044 | 4.64813 | 0.744732 | 0.136559 |
| ABA 1h | 23.6289 | 3.33157 | 1.0128 | 0.148085 |
| ACC 1h | 20.4011 | 9.24679 | 0.612435 | 0.285362 |
| BABA 1h | 28.8132 | 3.70847 | 1.1374 | 0.148457 |
| Chitin 1h | 23.2074 | 8.61212 | 0.859514 | 0.282129 |
| Epi 1h | 27.2088 | 3.4832 | 1.20869 | 0.155758 |
| SA 1h | 31.9622 | 5.08021 | 1.16505 | 0.195856 |
| Me-JA 1h | 7.90235 | 3.26466 | 0.357723 | 0.160761 |
| Control 6h | 20.247 | 7.91514 | 0.619092 | 0.307254 |
| ABA 6h | 206.016 | 30.4843 | 7.29122 | 0.715392 |
| ACC 6h | 24.9471 | 4.26875 | 0.825959 | 0.141976 |
| BABA 6h | 28.7879 | 3.97956 | 0.982461 | 0.136464 |
| Chitin 6h | 23.0681 | 5.20622 | 0.785153 | 0.192534 |
| Epi 6h | 23.6574 | 7.23125 | 0.736278 | 0.175044 |
| SA 6h | 28.6542 | 4.25045 | 1.09164 | 0.152274 |
| Me-JA 6h | 23.1875 | 11.54 | 0.642829 | 0.436493 |
Source Transcript PGSC0003DMT400066480 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G49630.1 | +2 | 1e-63 | 207 | 95/137 (69%) | amino acid permease 6 | chr5:20142681-20146441 REVERSE LENGTH=481 |
