Probe CUST_14434_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14434_PI426222305 | JHI_St_60k_v1 | DMT400066571 | CACAACATCATCATCTTCTTCTGTTTGAGCCAAAAATAAAAACACGAAGCCAATGTCTTT |
All Microarray Probes Designed to Gene DMG400025875
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14434_PI426222305 | JHI_St_60k_v1 | DMT400066571 | CACAACATCATCATCTTCTTCTGTTTGAGCCAAAAATAAAAACACGAAGCCAATGTCTTT |
Microarray Signals from CUST_14434_PI426222305
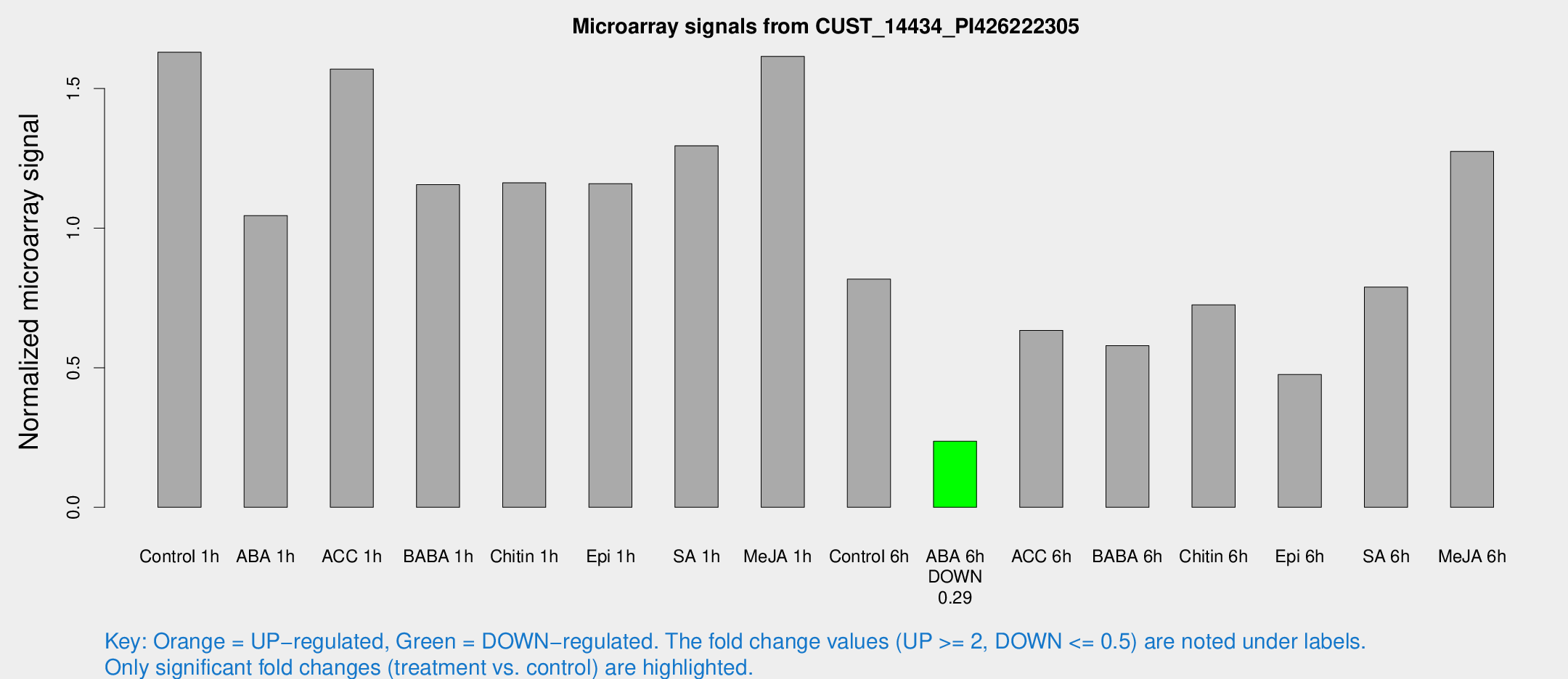
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1506.23 | 140.601 | 1.63024 | 0.0942076 |
| ABA 1h | 867.099 | 129.257 | 1.04521 | 0.14508 |
| ACC 1h | 1554.49 | 325.927 | 1.56979 | 0.247438 |
| BABA 1h | 1024.74 | 64.4415 | 1.15573 | 0.197184 |
| Chitin 1h | 982.115 | 161.688 | 1.16235 | 0.199423 |
| Epi 1h | 936.189 | 118.893 | 1.15938 | 0.14403 |
| SA 1h | 1296.07 | 315.341 | 1.29511 | 0.392337 |
| Me-JA 1h | 1215.03 | 89.4721 | 1.61541 | 0.225542 |
| Control 6h | 817.632 | 208.332 | 0.817398 | 0.18182 |
| ABA 6h | 236.242 | 36.9715 | 0.236824 | 0.0506687 |
| ACC 6h | 680.814 | 100.382 | 0.633967 | 0.0368471 |
| BABA 6h | 596.838 | 41.6535 | 0.579125 | 0.0450821 |
| Chitin 6h | 718.182 | 85.0111 | 0.725516 | 0.107672 |
| Epi 6h | 510.869 | 92.7316 | 0.475474 | 0.136986 |
| SA 6h | 720.759 | 62.7984 | 0.788755 | 0.153001 |
| Me-JA 6h | 1173.08 | 105.609 | 1.27517 | 0.073752 |
Source Transcript PGSC0003DMT400066571 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G67210.1 | +3 | 5e-106 | 322 | 170/304 (56%) | Protein of unknown function (DUF579) | chr5:26819019-26819972 FORWARD LENGTH=317 |
