Probe CUST_14272_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14272_PI426222305 | JHI_St_60k_v1 | DMT400060013 | CCCACATATTCGCATCAAGTAATAGGTCTTTCAACATTGGTTTTGGAATTCTTAGAAACT |
All Microarray Probes Designed to Gene DMG400023352
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14246_PI426222305 | JHI_St_60k_v1 | DMT400060014 | CCCACATATTCGCATCAAGTAATAGGTCTTTCAACATTGGTTTTGGAATTCTTAGAAACT |
| CUST_14272_PI426222305 | JHI_St_60k_v1 | DMT400060013 | CCCACATATTCGCATCAAGTAATAGGTCTTTCAACATTGGTTTTGGAATTCTTAGAAACT |
Microarray Signals from CUST_14272_PI426222305
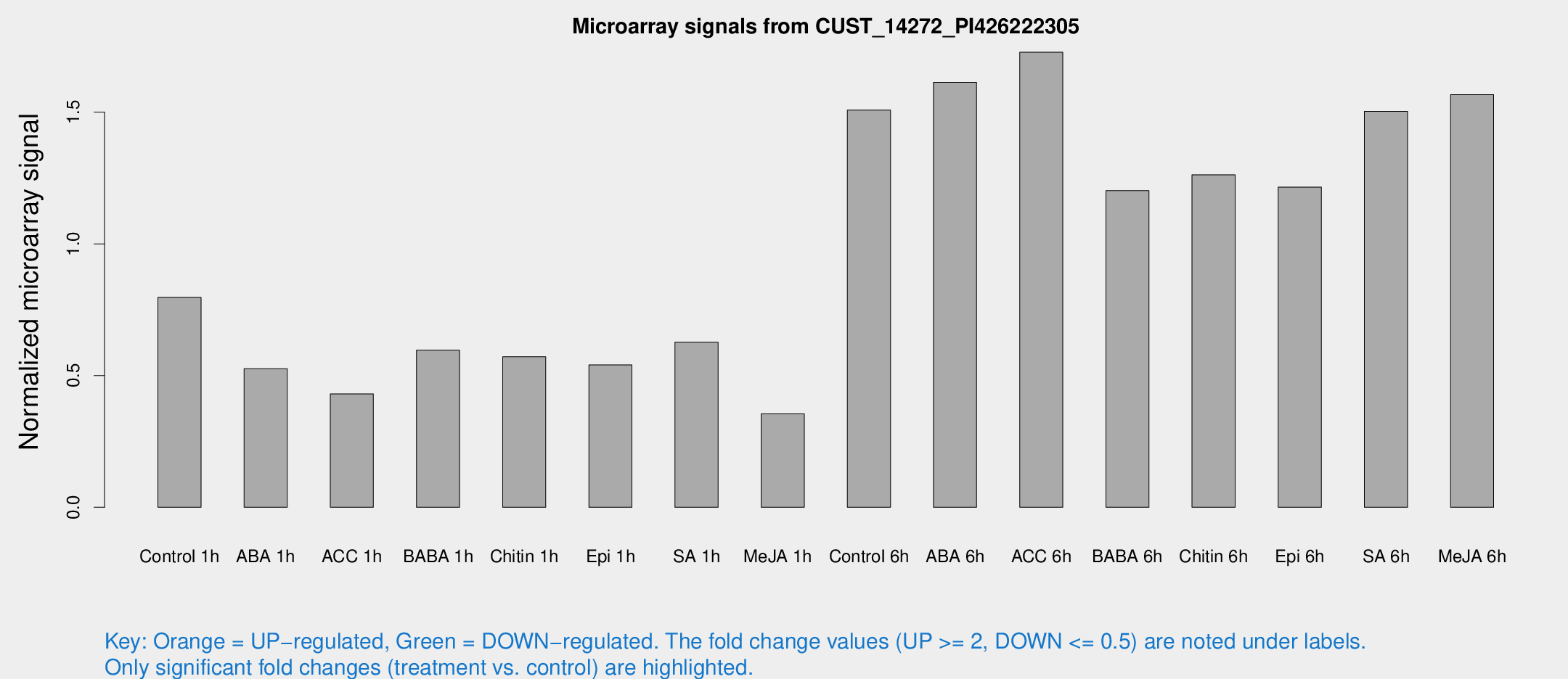
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1076.01 | 168.265 | 0.79629 | 0.0702918 |
| ABA 1h | 618.014 | 48.2613 | 0.525893 | 0.0485965 |
| ACC 1h | 644.445 | 174.339 | 0.430434 | 0.117132 |
| BABA 1h | 809.245 | 182.478 | 0.596309 | 0.0973099 |
| Chitin 1h | 689.516 | 89.851 | 0.571439 | 0.03416 |
| Epi 1h | 636.765 | 103.576 | 0.540255 | 0.0920104 |
| SA 1h | 865.847 | 118.394 | 0.626693 | 0.0461124 |
| Me-JA 1h | 393.051 | 67.0346 | 0.354635 | 0.0291356 |
| Control 6h | 2077.57 | 384.387 | 1.50758 | 0.180151 |
| ABA 6h | 2375.78 | 493.283 | 1.61281 | 0.282402 |
| ACC 6h | 2621.32 | 151.645 | 1.72726 | 0.199797 |
| BABA 6h | 1796.45 | 181.208 | 1.20191 | 0.087207 |
| Chitin 6h | 1779.25 | 103.068 | 1.26223 | 0.07292 |
| Epi 6h | 1824.76 | 159.186 | 1.21547 | 0.0702217 |
| SA 6h | 1989.41 | 220.189 | 1.50305 | 0.0868223 |
| Me-JA 6h | 2116.63 | 361.621 | 1.56654 | 0.155164 |
Source Transcript PGSC0003DMT400060013 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G04350.1 | +2 | 0.0 | 692 | 340/573 (59%) | Plant protein of unknown function (DUF946) | chr3:1153972-1156469 REVERSE LENGTH=567 |
