Probe CUST_14246_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14246_PI426222305 | JHI_St_60k_v1 | DMT400060014 | CCCACATATTCGCATCAAGTAATAGGTCTTTCAACATTGGTTTTGGAATTCTTAGAAACT |
All Microarray Probes Designed to Gene DMG400023352
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14272_PI426222305 | JHI_St_60k_v1 | DMT400060013 | CCCACATATTCGCATCAAGTAATAGGTCTTTCAACATTGGTTTTGGAATTCTTAGAAACT |
| CUST_14246_PI426222305 | JHI_St_60k_v1 | DMT400060014 | CCCACATATTCGCATCAAGTAATAGGTCTTTCAACATTGGTTTTGGAATTCTTAGAAACT |
Microarray Signals from CUST_14246_PI426222305
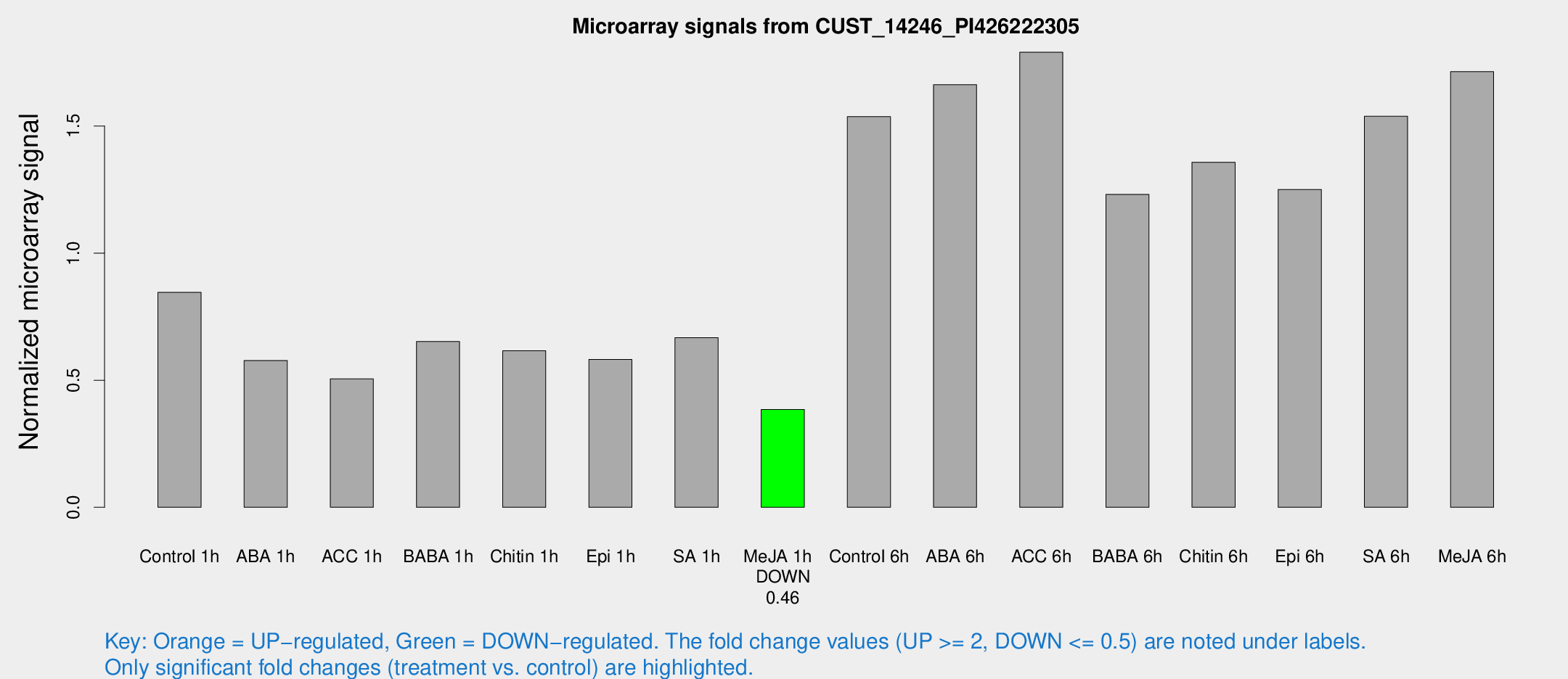
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1083.56 | 121.359 | 0.84578 | 0.0489102 |
| ABA 1h | 648.94 | 40.0707 | 0.577314 | 0.0503282 |
| ACC 1h | 713.623 | 179.394 | 0.505111 | 0.119909 |
| BABA 1h | 838.862 | 179.329 | 0.652067 | 0.0905546 |
| Chitin 1h | 710.688 | 84.3272 | 0.616045 | 0.0417775 |
| Epi 1h | 656.583 | 104.59 | 0.581908 | 0.0951712 |
| SA 1h | 876.438 | 86.8218 | 0.667118 | 0.0386072 |
| Me-JA 1h | 404.663 | 52.0034 | 0.385177 | 0.0225004 |
| Control 6h | 2044.19 | 405.143 | 1.53717 | 0.211015 |
| ABA 6h | 2322.31 | 422.876 | 1.66244 | 0.246382 |
| ACC 6h | 2607.43 | 151.006 | 1.7905 | 0.160521 |
| BABA 6h | 1771.48 | 218.524 | 1.23086 | 0.125654 |
| Chitin 6h | 1829.92 | 105.754 | 1.35745 | 0.0784281 |
| Epi 6h | 1807.37 | 191.621 | 1.2507 | 0.0722669 |
| SA 6h | 1962.54 | 260.067 | 1.53858 | 0.0888846 |
| Me-JA 6h | 2211.99 | 341.11 | 1.71389 | 0.122069 |
Source Transcript PGSC0003DMT400060014 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G04350.1 | +2 | 0.0 | 655 | 324/536 (60%) | Plant protein of unknown function (DUF946) | chr3:1153972-1156469 REVERSE LENGTH=567 |
