Probe CUST_14221_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14221_PI426222305 | JHI_St_60k_v1 | DMT400060061 | GTGGAACTTCACTCTTTGCCTATTGTGTATATTGAAATGCCAAATGATATGTCTCACTTA |
All Microarray Probes Designed to Gene DMG400023366
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14221_PI426222305 | JHI_St_60k_v1 | DMT400060061 | GTGGAACTTCACTCTTTGCCTATTGTGTATATTGAAATGCCAAATGATATGTCTCACTTA |
| CUST_14107_PI426222305 | JHI_St_60k_v1 | DMT400060058 | AAGGAAGAAGGAAGACATTGTAGCAGCTGTGCAGAAGAGTGGAATTGCTTGGGCATTGAA |
Microarray Signals from CUST_14221_PI426222305
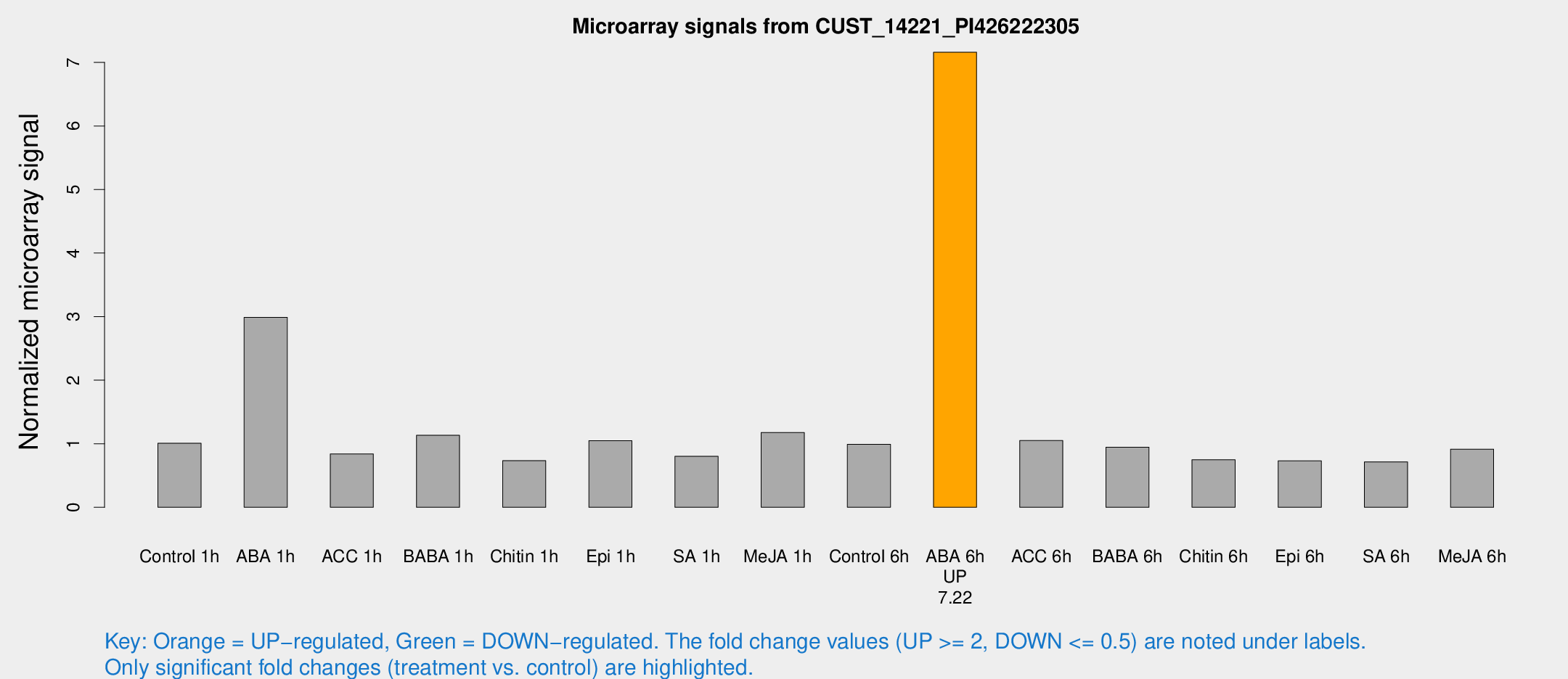
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 10.953 | 3.30803 | 1.00741 | 0.375674 |
| ABA 1h | 28.5921 | 7.48737 | 2.9878 | 0.843993 |
| ACC 1h | 8.81829 | 4.10023 | 0.839307 | 0.409702 |
| BABA 1h | 11.357 | 3.53352 | 1.13271 | 0.399308 |
| Chitin 1h | 6.58193 | 3.24488 | 0.733269 | 0.36395 |
| Epi 1h | 11.1573 | 5.45377 | 1.04849 | 0.570992 |
| SA 1h | 8.55474 | 3.33618 | 0.802288 | 0.343247 |
| Me-JA 1h | 9.50407 | 3.3985 | 1.17697 | 0.418513 |
| Control 6h | 10.4172 | 3.40183 | 0.991679 | 0.37843 |
| ABA 6h | 75.5505 | 5.6798 | 7.15998 | 0.529929 |
| ACC 6h | 12.0109 | 4.03361 | 1.04955 | 0.341127 |
| BABA 6h | 12.1803 | 4.91373 | 0.946192 | 0.39218 |
| Chitin 6h | 8.14393 | 3.73489 | 0.747489 | 0.371176 |
| Epi 6h | 8.34373 | 3.97388 | 0.730001 | 0.356508 |
| SA 6h | 7.02453 | 3.60752 | 0.715048 | 0.37095 |
| Me-JA 6h | 9.40759 | 3.38788 | 0.914873 | 0.369302 |
Source Transcript PGSC0003DMT400060061 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
