Probe CUST_14079_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14079_PI426222305 | JHI_St_60k_v1 | DMT400060243 | GCAAATACACAATGATTGAAGGGGATCCATTGGGACAAAATCTTGATTTTATTTCTCATG |
All Microarray Probes Designed to Gene DMG400023435
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14079_PI426222305 | JHI_St_60k_v1 | DMT400060243 | GCAAATACACAATGATTGAAGGGGATCCATTGGGACAAAATCTTGATTTTATTTCTCATG |
Microarray Signals from CUST_14079_PI426222305
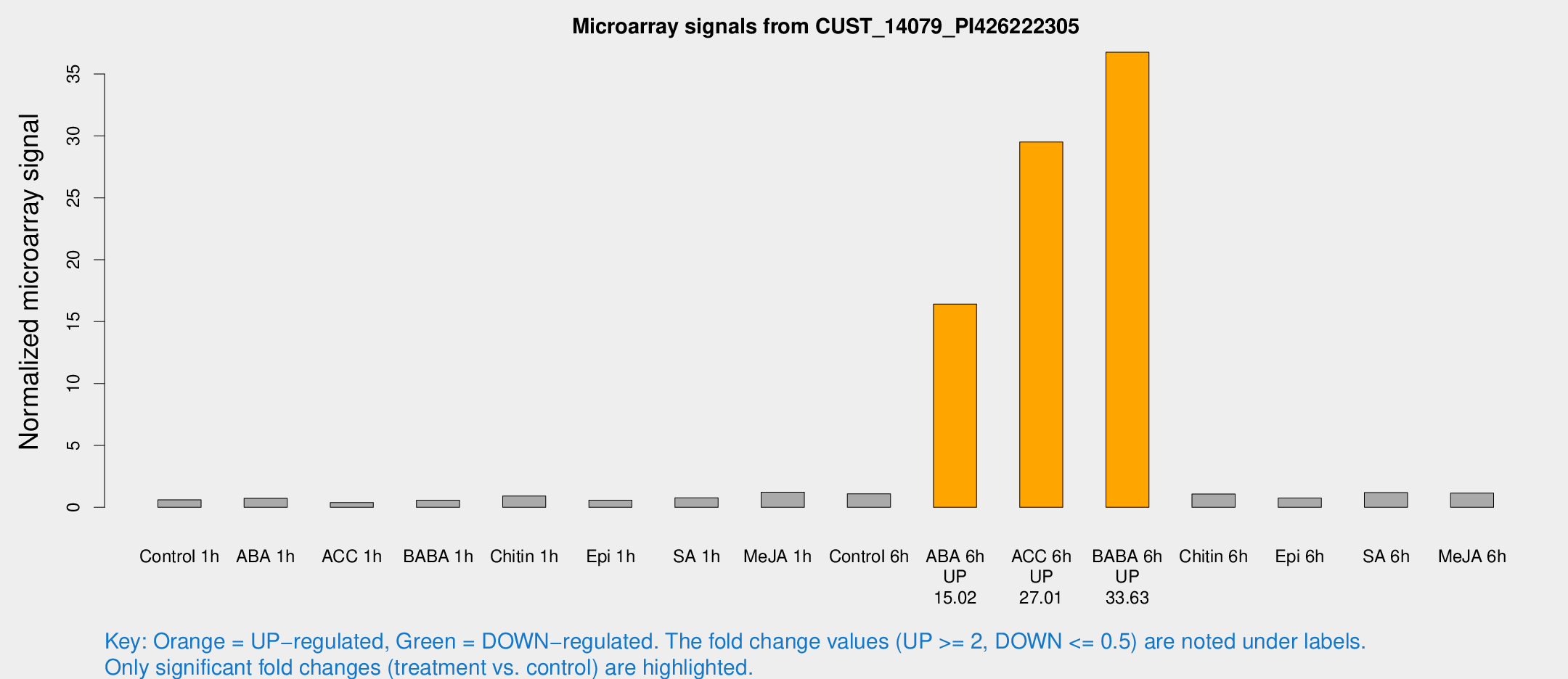
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.2998 | 6.38234 | 0.599893 | 0.326783 |
| ABA 1h | 13.5192 | 8.58391 | 0.729723 | 0.605458 |
| ACC 1h | 5.63551 | 3.27944 | 0.384284 | 0.222599 |
| BABA 1h | 8.8788 | 3.27392 | 0.57214 | 0.258779 |
| Chitin 1h | 12.8308 | 4.33058 | 0.905877 | 0.252415 |
| Epi 1h | 7.81084 | 2.91319 | 0.581567 | 0.264692 |
| SA 1h | 12.8228 | 4.11876 | 0.761984 | 0.28966 |
| Me-JA 1h | 18.5153 | 8.28517 | 1.2274 | 0.690219 |
| Control 6h | 22.7002 | 12.8819 | 1.09291 | 0.804033 |
| ABA 6h | 274.914 | 78.0946 | 16.4107 | 5.04019 |
| ACC 6h | 492.126 | 76.0531 | 29.5151 | 7.86683 |
| BABA 6h | 663.544 | 211.9 | 36.7598 | 15.2159 |
| Chitin 6h | 17.0727 | 3.80889 | 1.07303 | 0.243721 |
| Epi 6h | 14.2746 | 5.24328 | 0.749769 | 0.326609 |
| SA 6h | 18.3955 | 5.76382 | 1.1864 | 0.581711 |
| Me-JA 6h | 20.0581 | 7.21163 | 1.14032 | 0.772642 |
Source Transcript PGSC0003DMT400060243 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G45860.1 | +2 | 7e-09 | 54 | 26/59 (44%) | PYR1-like 11 | chr5:18604101-18604586 REVERSE LENGTH=161 |
