Probe CUST_13599_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13599_PI426222305 | JHI_St_60k_v1 | DMT400017544 | TCCCTTGGAGTTTAGATGCGTCTATTCATGATATTCCATTTTCGAAGGATTTGTTGACGT |
All Microarray Probes Designed to Gene DMG400006802
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13507_PI426222305 | JHI_St_60k_v1 | DMT400017543 | GCACCAATGTGACAATTTTGGCAGTTTCAAAGAGGATGTTTTTATTACTAAGTAGATGTG |
| CUST_13599_PI426222305 | JHI_St_60k_v1 | DMT400017544 | TCCCTTGGAGTTTAGATGCGTCTATTCATGATATTCCATTTTCGAAGGATTTGTTGACGT |
Microarray Signals from CUST_13599_PI426222305
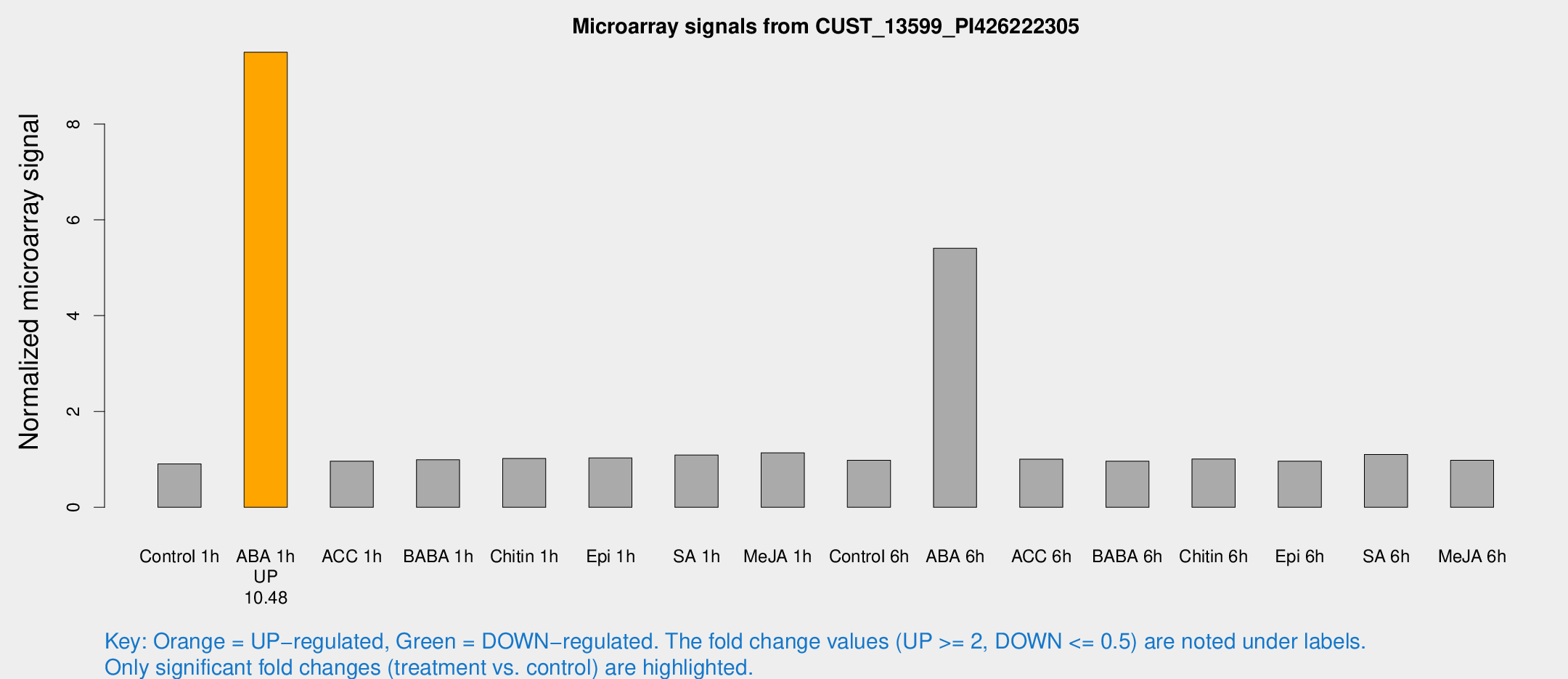
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.61873 | 3.25751 | 0.905844 | 0.524531 |
| ABA 1h | 53.1528 | 7.26855 | 9.49701 | 0.940385 |
| ACC 1h | 6.13468 | 3.55348 | 0.963959 | 0.558204 |
| BABA 1h | 5.93475 | 3.44477 | 0.99154 | 0.574157 |
| Chitin 1h | 5.70311 | 3.32559 | 1.01916 | 0.588786 |
| Epi 1h | 5.53977 | 3.21525 | 1.03071 | 0.596855 |
| SA 1h | 7.28523 | 3.2331 | 1.08995 | 0.534604 |
| Me-JA 1h | 5.78741 | 3.36885 | 1.13844 | 0.65917 |
| Control 6h | 6.10911 | 3.54313 | 0.98275 | 0.569319 |
| ABA 6h | 37.2557 | 7.54924 | 5.40687 | 0.902165 |
| ACC 6h | 7.2557 | 4.16191 | 1.0051 | 0.56215 |
| BABA 6h | 6.70414 | 3.89851 | 0.962825 | 0.557554 |
| Chitin 6h | 6.65524 | 3.85623 | 1.00848 | 0.583935 |
| Epi 6h | 6.81006 | 3.99305 | 0.963148 | 0.557717 |
| SA 6h | 6.79913 | 3.67068 | 1.10437 | 0.601672 |
| Me-JA 6h | 6.05976 | 3.51519 | 0.980551 | 0.567797 |
Source Transcript PGSC0003DMT400017544 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G07985.1 | +3 | 2e-07 | 50 | 32/63 (51%) | Expressed protein | chr1:2475508-2475942 FORWARD LENGTH=144 |
