Probe CUST_13564_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13564_PI426222305 | JHI_St_60k_v1 | DMT400017573 | CTGGAAATTCTACTCTTTGACCTGTTATTGCAACCTTAATTTTAGTTTGCAGAGAAGTGT |
All Microarray Probes Designed to Gene DMG400006816
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13564_PI426222305 | JHI_St_60k_v1 | DMT400017573 | CTGGAAATTCTACTCTTTGACCTGTTATTGCAACCTTAATTTTAGTTTGCAGAGAAGTGT |
Microarray Signals from CUST_13564_PI426222305
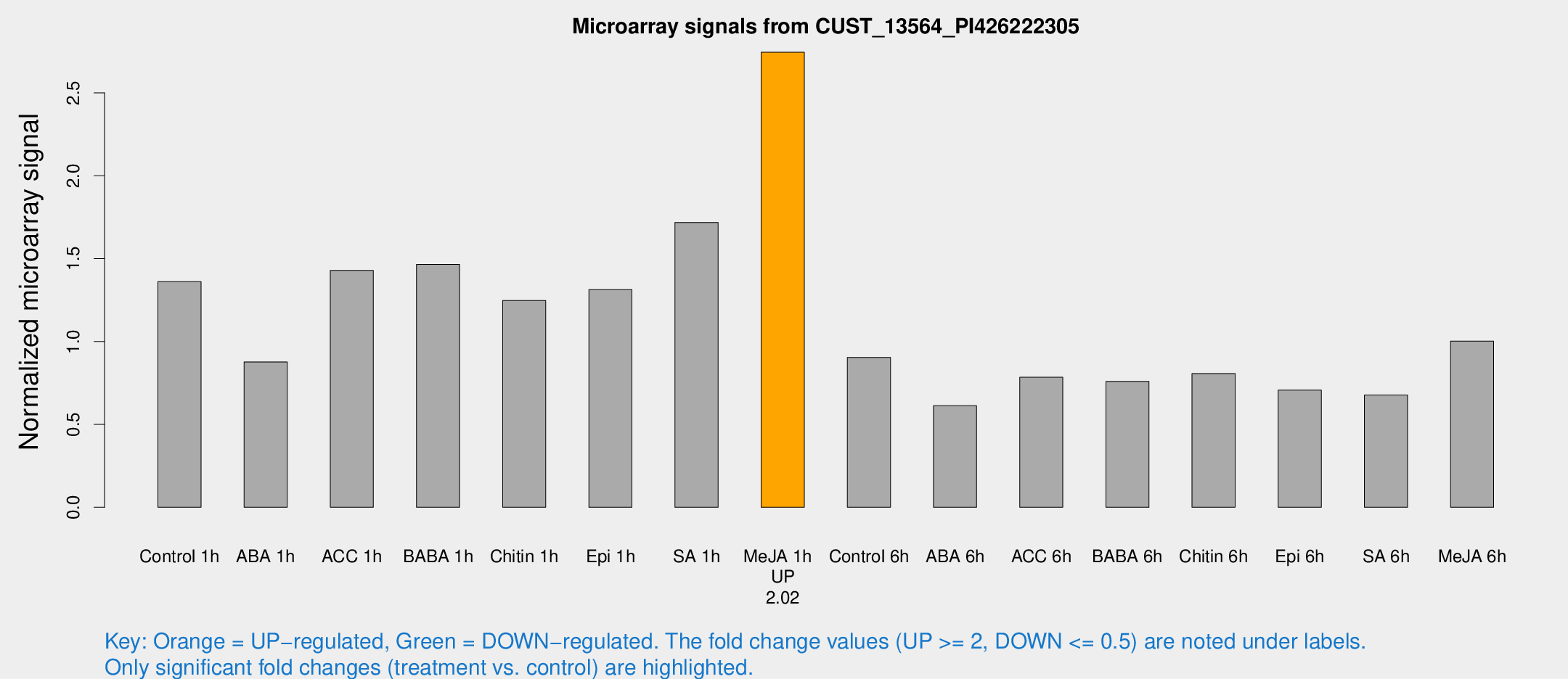
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1136.91 | 138.704 | 1.36105 | 0.115115 |
| ABA 1h | 643.776 | 57.6016 | 0.877031 | 0.0508054 |
| ACC 1h | 1257.64 | 244.8 | 1.42922 | 0.204866 |
| BABA 1h | 1175.49 | 115.65 | 1.46553 | 0.0847153 |
| Chitin 1h | 943.841 | 141.769 | 1.24785 | 0.190801 |
| Epi 1h | 941.897 | 71.5497 | 1.31351 | 0.0934279 |
| SA 1h | 1566.97 | 444.808 | 1.71826 | 0.527859 |
| Me-JA 1h | 1850.24 | 107.113 | 2.74563 | 0.15859 |
| Control 6h | 809.059 | 200.528 | 0.904344 | 0.193061 |
| ABA 6h | 541.596 | 53.9799 | 0.612689 | 0.0420877 |
| ACC 6h | 748.828 | 76.3935 | 0.784878 | 0.0454936 |
| BABA 6h | 712.979 | 91.6088 | 0.759318 | 0.0771042 |
| Chitin 6h | 708.253 | 41.0787 | 0.807266 | 0.046783 |
| Epi 6h | 671.821 | 105.547 | 0.706677 | 0.13179 |
| SA 6h | 567.872 | 96.7117 | 0.677638 | 0.0713803 |
| Me-JA 6h | 827.544 | 63.7453 | 1.00293 | 0.0589 |
Source Transcript PGSC0003DMT400017573 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G63940.1 | +3 | 0.0 | 609 | 341/674 (51%) | Protein kinase protein with adenine nucleotide alpha hydrolases-like domain | chr5:25588254-25591229 FORWARD LENGTH=705 |
