Probe CUST_13511_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13511_PI426222305 | JHI_St_60k_v1 | DMT400017664 | GAGAGAAGGATATAGGGTAGTTTCCAGGATATTCTTCATTGCAGATAACATACAATTGTT |
All Microarray Probes Designed to Gene DMG400006854
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13586_PI426222305 | JHI_St_60k_v1 | DMT400017667 | GAGAGAAGGATATAGGGTAGTTTCCAGGATATTCTTCATTGCAGATAACATACAATTGTT |
| CUST_13511_PI426222305 | JHI_St_60k_v1 | DMT400017664 | GAGAGAAGGATATAGGGTAGTTTCCAGGATATTCTTCATTGCAGATAACATACAATTGTT |
Microarray Signals from CUST_13511_PI426222305
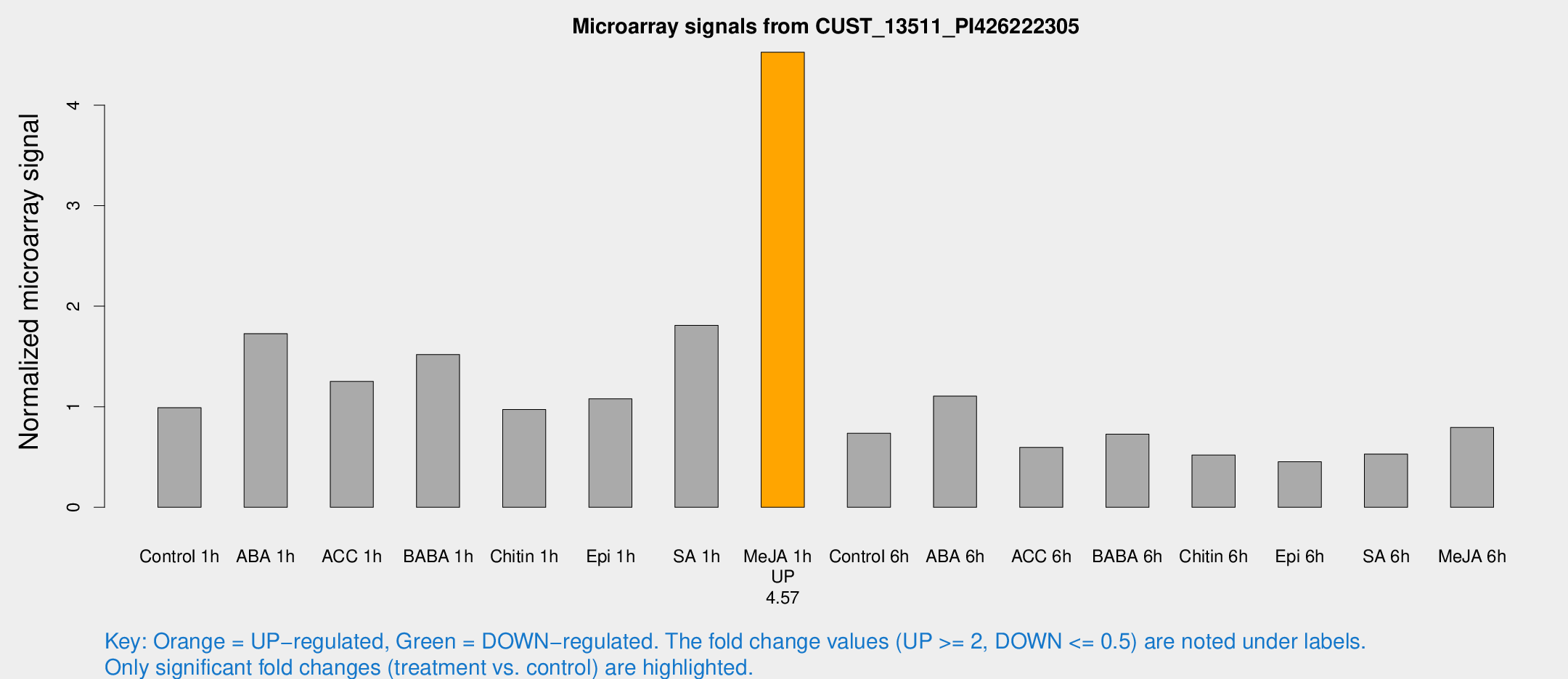
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 440.385 | 87.6566 | 0.990728 | 0.249028 |
| ABA 1h | 692.595 | 186.951 | 1.72751 | 0.331273 |
| ACC 1h | 584.713 | 152.418 | 1.25176 | 0.27159 |
| BABA 1h | 624.313 | 48.463 | 1.51977 | 0.0983161 |
| Chitin 1h | 374.554 | 40.502 | 0.972212 | 0.147035 |
| Epi 1h | 401.295 | 45.0195 | 1.0794 | 0.11455 |
| SA 1h | 942.445 | 418.353 | 1.80989 | 0.846494 |
| Me-JA 1h | 1580.07 | 152.968 | 4.52608 | 0.261521 |
| Control 6h | 358.349 | 109.15 | 0.736955 | 0.241613 |
| ABA 6h | 499.439 | 29.1651 | 1.1058 | 0.064405 |
| ACC 6h | 290.19 | 17.3448 | 0.596399 | 0.0942005 |
| BABA 6h | 346.922 | 27.2509 | 0.726786 | 0.0428801 |
| Chitin 6h | 235.475 | 14.2428 | 0.520286 | 0.0313904 |
| Epi 6h | 221.76 | 35.6789 | 0.453019 | 0.0670074 |
| SA 6h | 222.482 | 13.4604 | 0.529409 | 0.067392 |
| Me-JA 6h | 342.615 | 47.1321 | 0.795012 | 0.0661708 |
Source Transcript PGSC0003DMT400017664 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G13700.1 | +1 | 3e-71 | 231 | 115/164 (70%) | polyamine oxidase 1 | chr5:4420222-4422974 REVERSE LENGTH=472 |
