Probe CUST_13483_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13483_PI426222305 | JHI_St_60k_v1 | DMT400017518 | CAAAACCAGCATCAAGTGCATGTATTGCAAATGACTCAATACAGTAACTGCATTTGTAAA |
All Microarray Probes Designed to Gene DMG400006797
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13483_PI426222305 | JHI_St_60k_v1 | DMT400017518 | CAAAACCAGCATCAAGTGCATGTATTGCAAATGACTCAATACAGTAACTGCATTTGTAAA |
Microarray Signals from CUST_13483_PI426222305
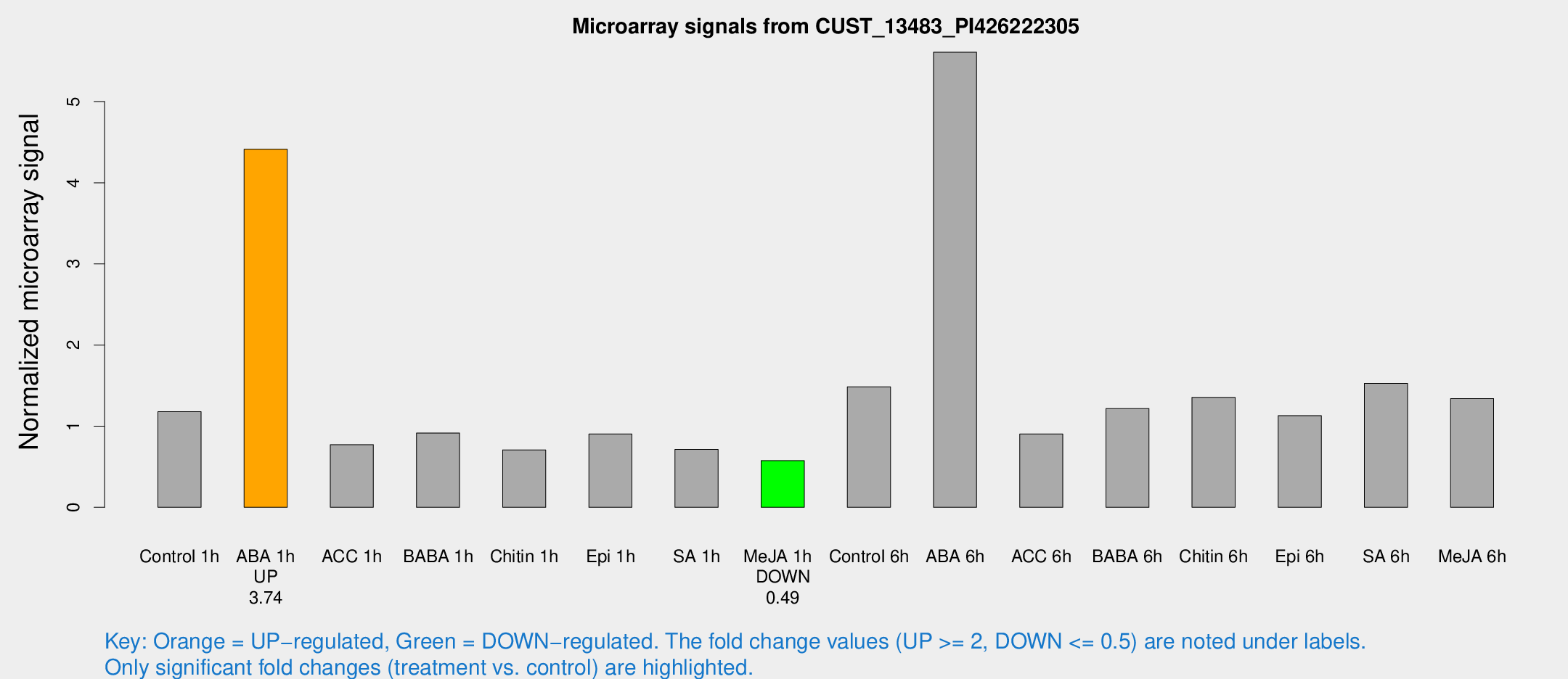
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 232.011 | 16.387 | 1.17915 | 0.0699498 |
| ABA 1h | 769.906 | 64.4213 | 4.4136 | 0.311703 |
| ACC 1h | 160.383 | 29.1213 | 0.771951 | 0.0897378 |
| BABA 1h | 178.929 | 31.0469 | 0.915208 | 0.081642 |
| Chitin 1h | 126.592 | 16.598 | 0.707318 | 0.044772 |
| Epi 1h | 155.791 | 21.1766 | 0.903063 | 0.12372 |
| SA 1h | 145.696 | 17.8338 | 0.714303 | 0.0991616 |
| Me-JA 1h | 93.8629 | 13.4146 | 0.576195 | 0.0455452 |
| Control 6h | 316.117 | 89.4866 | 1.48581 | 0.343637 |
| ABA 6h | 1242.82 | 277.628 | 5.60911 | 1.28929 |
| ACC 6h | 205.162 | 22.2353 | 0.903397 | 0.0549773 |
| BABA 6h | 285.359 | 77.0961 | 1.21762 | 0.305197 |
| Chitin 6h | 285.84 | 32.1237 | 1.35532 | 0.11295 |
| Epi 6h | 261.112 | 58.171 | 1.13047 | 0.241222 |
| SA 6h | 333.255 | 100.374 | 1.52657 | 0.736252 |
| Me-JA 6h | 281.669 | 68.3345 | 1.33998 | 0.29135 |
Source Transcript PGSC0003DMT400017518 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G64290.1 | +2 | 4e-83 | 282 | 147/205 (72%) | dicarboxylate transport 2.1 | chr5:25714495-25716642 REVERSE LENGTH=563 |
