Probe CUST_13205_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13205_PI426222305 | JHI_St_60k_v1 | DMT400007511 | GAAGAAGAGAATTATTTGAACTACGATGGATTTCAATCACAAGGGACTTCATTTCTTTGG |
All Microarray Probes Designed to Gene DMG400002899
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13205_PI426222305 | JHI_St_60k_v1 | DMT400007511 | GAAGAAGAGAATTATTTGAACTACGATGGATTTCAATCACAAGGGACTTCATTTCTTTGG |
Microarray Signals from CUST_13205_PI426222305
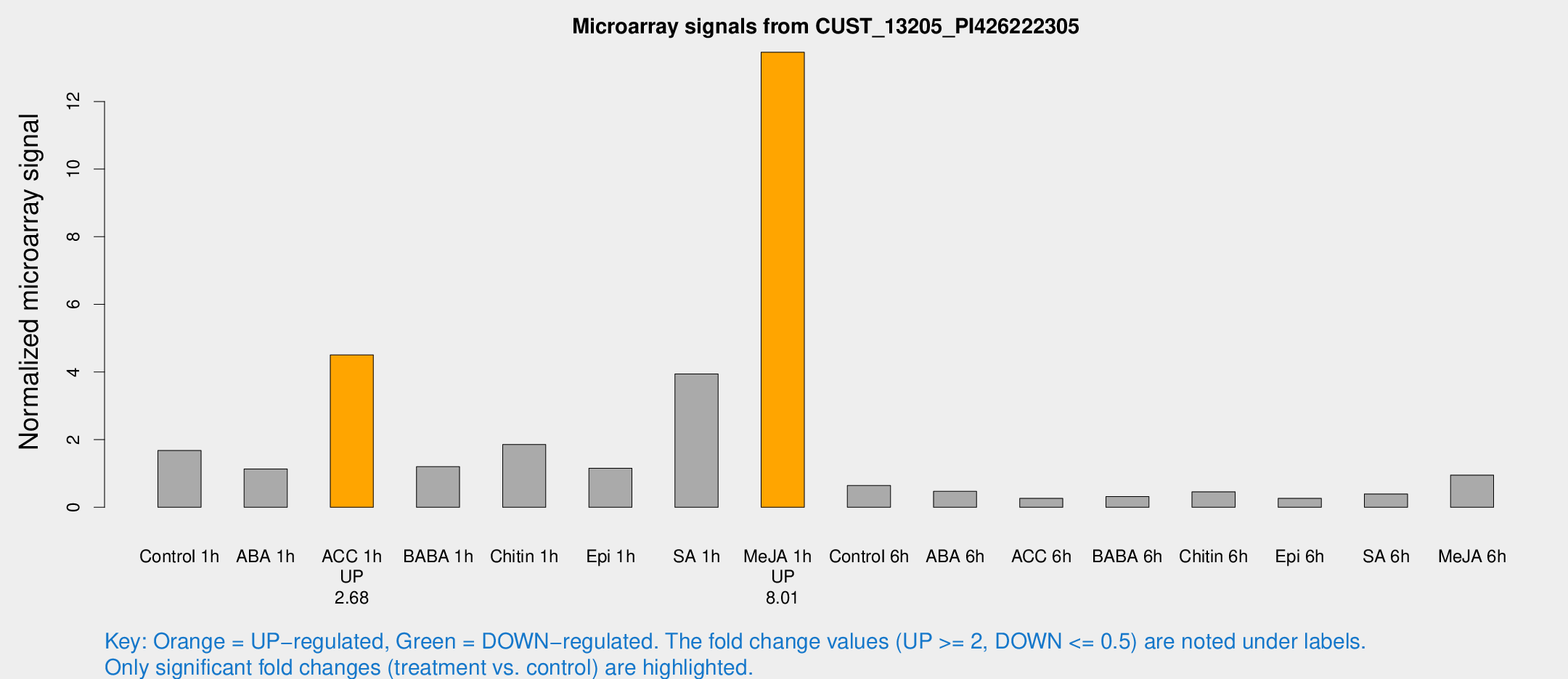
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 88.9288 | 22.8248 | 1.67894 | 0.465291 |
| ABA 1h | 50.0779 | 6.78957 | 1.13172 | 0.0936634 |
| ACC 1h | 241.779 | 53.2232 | 4.50649 | 0.83447 |
| BABA 1h | 61.6824 | 15.9776 | 1.20101 | 0.240753 |
| Chitin 1h | 82.6589 | 6.75376 | 1.85753 | 0.194492 |
| Epi 1h | 50.2593 | 6.81202 | 1.15682 | 0.153876 |
| SA 1h | 200.989 | 19.3575 | 3.94296 | 0.356384 |
| Me-JA 1h | 553.765 | 81.859 | 13.456 | 1.26755 |
| Control 6h | 41.0539 | 21.0575 | 0.645717 | 0.311864 |
| ABA 6h | 26.995 | 7.03199 | 0.471484 | 0.139048 |
| ACC 6h | 17.1526 | 6.51633 | 0.265144 | 0.071524 |
| BABA 6h | 20.2017 | 7.40064 | 0.317172 | 0.134973 |
| Chitin 6h | 28.3279 | 9.2629 | 0.460072 | 0.239291 |
| Epi 6h | 16.4182 | 4.64026 | 0.265217 | 0.129641 |
| SA 6h | 19.7349 | 3.48199 | 0.395705 | 0.0753593 |
| Me-JA 6h | 47.4153 | 6.07712 | 0.953306 | 0.0916495 |
Source Transcript PGSC0003DMT400007511 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G19210.1 | +2 | 6e-48 | 162 | 89/190 (47%) | Integrase-type DNA-binding superfamily protein | chr1:6626973-6627530 REVERSE LENGTH=185 |
