Probe CUST_12938_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12938_PI426222305 | JHI_St_60k_v1 | DMT400063172 | CTGTTTTGTACAGACCAGAAATATGTGCCTTACTGTACCTGTTTTATGCCTTTACAAATA |
All Microarray Probes Designed to Gene DMG400024572
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12938_PI426222305 | JHI_St_60k_v1 | DMT400063172 | CTGTTTTGTACAGACCAGAAATATGTGCCTTACTGTACCTGTTTTATGCCTTTACAAATA |
Microarray Signals from CUST_12938_PI426222305
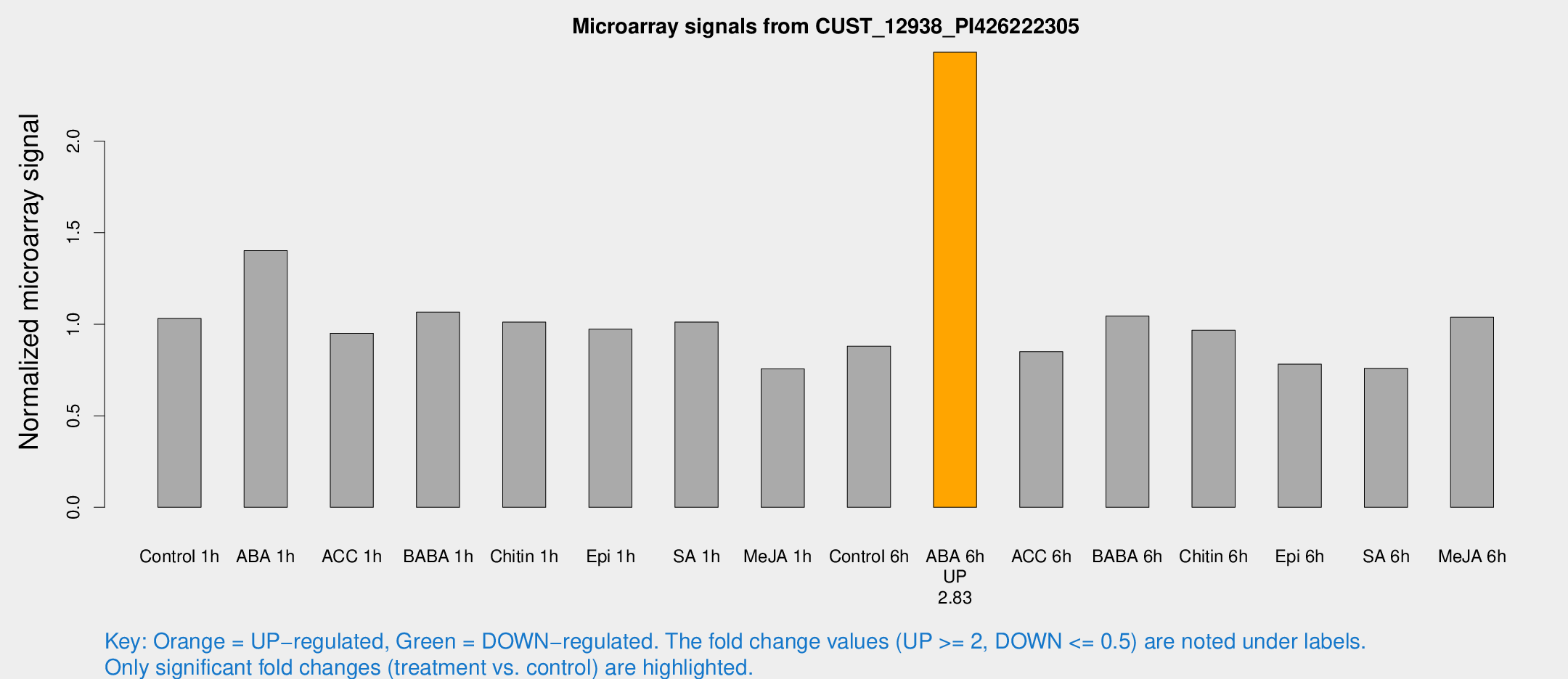
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9215.03 | 579.013 | 1.03135 | 0.0595461 |
| ABA 1h | 11421 | 2026.67 | 1.4016 | 0.173273 |
| ACC 1h | 9463.37 | 2401.05 | 0.950647 | 0.229552 |
| BABA 1h | 9651.34 | 2025.23 | 1.0663 | 0.147362 |
| Chitin 1h | 8234.89 | 1116.75 | 1.01136 | 0.0697215 |
| Epi 1h | 7663.91 | 1084.62 | 0.972609 | 0.135446 |
| SA 1h | 9342.35 | 942.588 | 1.01188 | 0.0650931 |
| Me-JA 1h | 5581.61 | 714.628 | 0.75641 | 0.0488355 |
| Control 6h | 8239.54 | 1848.95 | 0.879571 | 0.151275 |
| ABA 6h | 23572.1 | 1363.77 | 2.48568 | 0.143511 |
| ACC 6h | 8833.33 | 1164.77 | 0.850179 | 0.0490862 |
| BABA 6h | 10480 | 881.67 | 1.04396 | 0.0602737 |
| Chitin 6h | 9162.27 | 529.071 | 0.966846 | 0.055822 |
| Epi 6h | 7941.49 | 887.704 | 0.781688 | 0.045132 |
| SA 6h | 7000.68 | 1385.36 | 0.758717 | 0.0826496 |
| Me-JA 6h | 9714.12 | 2158.21 | 1.0381 | 0.161325 |
Source Transcript PGSC0003DMT400063172 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G19000.1 | +2 | 2e-48 | 169 | 135/287 (47%) | Homeodomain-like superfamily protein | chr1:6561335-6562684 REVERSE LENGTH=285 |
