Probe CUST_12936_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12936_PI426222305 | JHI_St_60k_v1 | DMT400062981 | ATTCCATCAGAGTTAGTCTACACCATGCAATAGATGTCACTTTTGTGCTAAATGCCTCTC |
All Microarray Probes Designed to Gene DMG400024508
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12936_PI426222305 | JHI_St_60k_v1 | DMT400062981 | ATTCCATCAGAGTTAGTCTACACCATGCAATAGATGTCACTTTTGTGCTAAATGCCTCTC |
Microarray Signals from CUST_12936_PI426222305
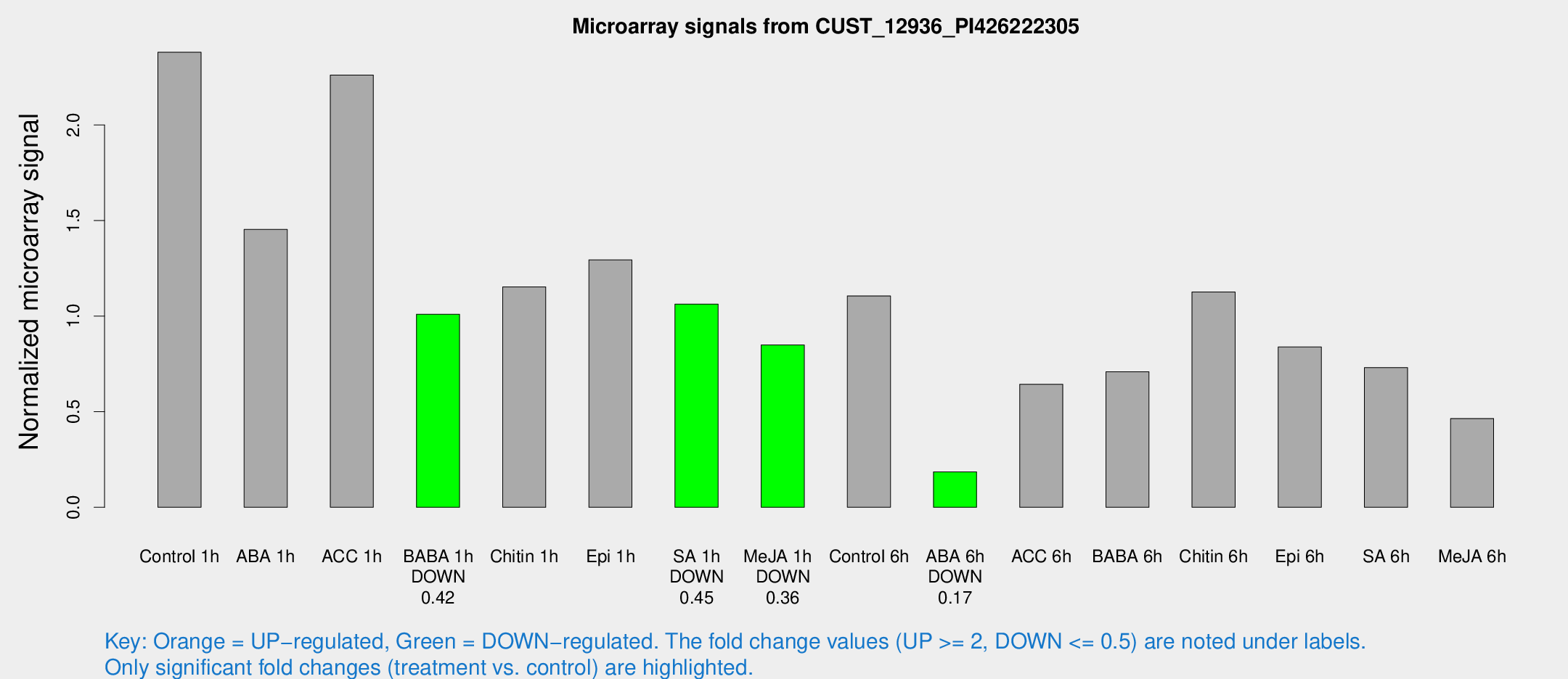
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3334.83 | 281.393 | 2.3803 | 0.137447 |
| ABA 1h | 1817.11 | 228.827 | 1.45381 | 0.290987 |
| ACC 1h | 3272.59 | 376.277 | 2.26103 | 0.432989 |
| BABA 1h | 1374.5 | 162.287 | 1.00988 | 0.0583632 |
| Chitin 1h | 1511.38 | 340.864 | 1.15312 | 0.255478 |
| Epi 1h | 1560.78 | 90.325 | 1.29421 | 0.0747686 |
| SA 1h | 1536.36 | 165.718 | 1.06297 | 0.104543 |
| Me-JA 1h | 993.496 | 180.072 | 0.84882 | 0.170254 |
| Control 6h | 1553.46 | 133.932 | 1.10584 | 0.0638934 |
| ABA 6h | 275.5 | 16.3323 | 0.185672 | 0.0167564 |
| ACC 6h | 1059.26 | 180.806 | 0.643786 | 0.053884 |
| BABA 6h | 1176.37 | 281.943 | 0.709245 | 0.208568 |
| Chitin 6h | 1669.38 | 96.5701 | 1.12607 | 0.0650639 |
| Epi 6h | 1316.9 | 76.213 | 0.838765 | 0.0771803 |
| SA 6h | 1013.77 | 87.9472 | 0.730945 | 0.0422821 |
| Me-JA 6h | 728.315 | 267.361 | 0.464206 | 0.131422 |
Source Transcript PGSC0003DMT400062981 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G23060.1 | +2 | 3e-160 | 466 | 236/357 (66%) | calcium sensing receptor | chr5:7736760-7738412 REVERSE LENGTH=387 |
