Probe CUST_12445_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12445_PI426222305 | JHI_St_60k_v1 | DMT400085559 | AAGCCATTGCAGAATCCTTGTCCAATGCAGCCTATTGCTTCTTCAGGATTGTTTAAATTT |
All Microarray Probes Designed to Gene DMG400035130
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12445_PI426222305 | JHI_St_60k_v1 | DMT400085559 | AAGCCATTGCAGAATCCTTGTCCAATGCAGCCTATTGCTTCTTCAGGATTGTTTAAATTT |
Microarray Signals from CUST_12445_PI426222305
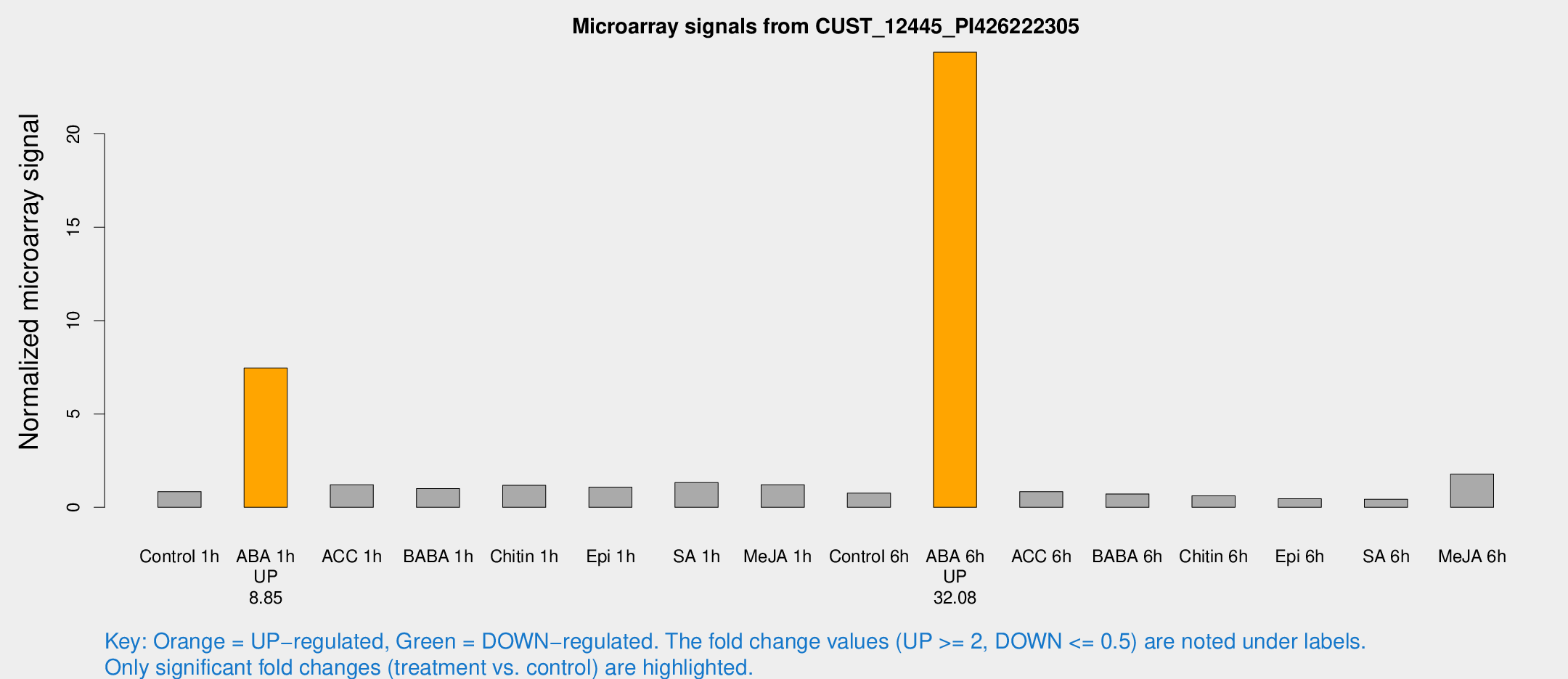
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 45.4234 | 7.38268 | 0.842579 | 0.167507 |
| ABA 1h | 348.595 | 34.2334 | 7.45981 | 0.997095 |
| ACC 1h | 67.4389 | 12.3008 | 1.20737 | 0.157101 |
| BABA 1h | 50.8666 | 4.58538 | 1.00593 | 0.116746 |
| Chitin 1h | 55.5588 | 4.59795 | 1.17487 | 0.0968742 |
| Epi 1h | 49.6112 | 6.70246 | 1.07747 | 0.159708 |
| SA 1h | 75.8695 | 19.9322 | 1.32674 | 0.337982 |
| Me-JA 1h | 56.8166 | 18.3626 | 1.21105 | 0.393302 |
| Control 6h | 44.7012 | 12.9214 | 0.759766 | 0.216888 |
| ABA 6h | 1437.92 | 314.421 | 24.3732 | 5.41829 |
| ACC 6h | 55.0771 | 15.9883 | 0.836499 | 0.321196 |
| BABA 6h | 42.0262 | 4.52682 | 0.715121 | 0.0771977 |
| Chitin 6h | 34.2303 | 4.27228 | 0.611044 | 0.0776739 |
| Epi 6h | 28.0123 | 4.64745 | 0.460685 | 0.114649 |
| SA 6h | 25.4849 | 7.86616 | 0.428915 | 0.130286 |
| Me-JA 6h | 97.2296 | 20.5421 | 1.78348 | 0.519827 |
Source Transcript PGSC0003DMT400085559 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G62420.1 | +1 | 3e-33 | 116 | 64/124 (52%) | basic region/leucine zipper motif 53 | chr3:23091844-23092284 REVERSE LENGTH=146 |
