Probe CUST_12409_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12409_PI426222305 | JHI_St_60k_v1 | DMT400002999 | GTTTTGTTTACAGGTAGTGAAAACTCTACAAAGTCTATTGCCAATCCCCGCGAACCAAAA |
All Microarray Probes Designed to Gene DMG400001181
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12384_PI426222305 | JHI_St_60k_v1 | DMT400003000 | ATTCTAGATGATTCTGGAACTGTGCTTGTTCTGGGAAACGTTGTGTTCCTCAGGCCTGAT |
| CUST_12409_PI426222305 | JHI_St_60k_v1 | DMT400002999 | GTTTTGTTTACAGGTAGTGAAAACTCTACAAAGTCTATTGCCAATCCCCGCGAACCAAAA |
Microarray Signals from CUST_12409_PI426222305
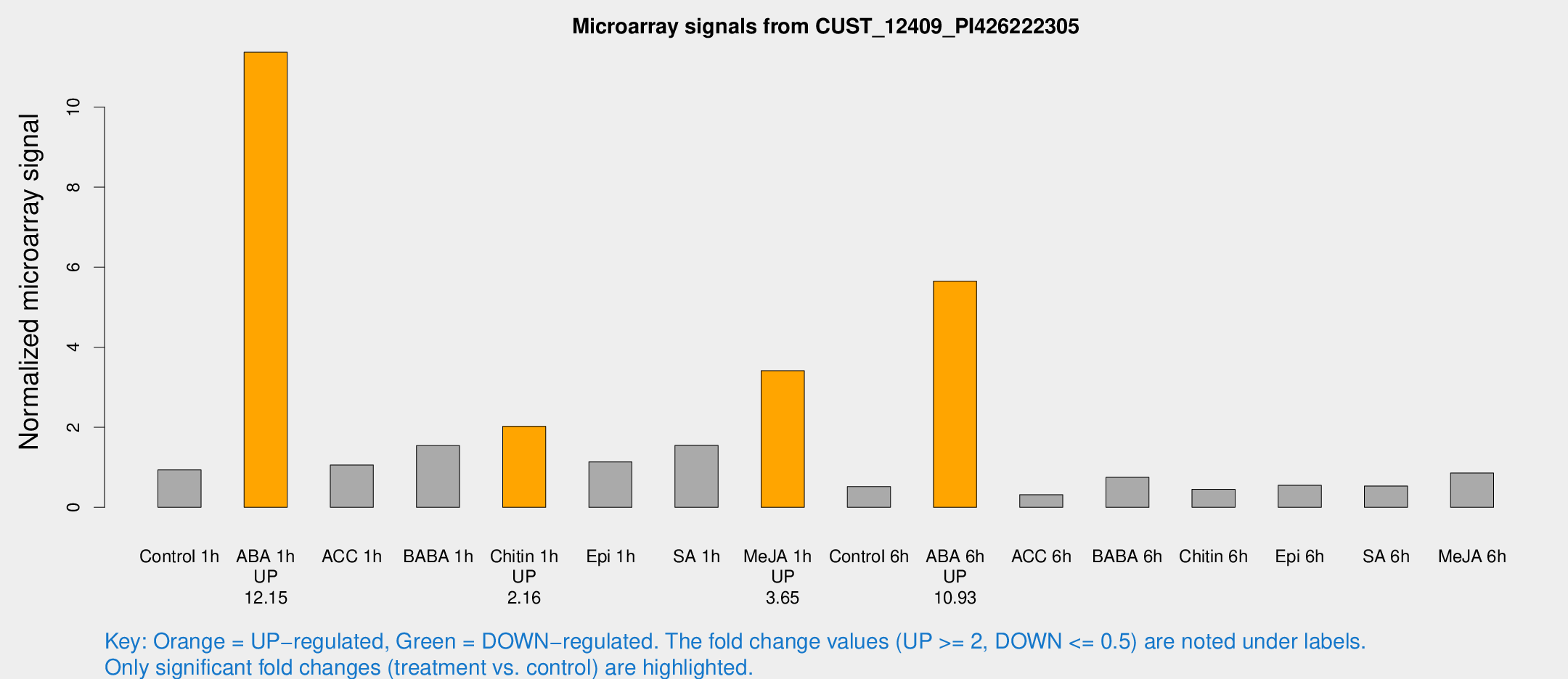
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 39.0761 | 3.90236 | 0.935756 | 0.108672 |
| ABA 1h | 426.724 | 61.3811 | 11.3712 | 1.04099 |
| ACC 1h | 46.698 | 9.24322 | 1.05592 | 0.141581 |
| BABA 1h | 67.5587 | 19.4848 | 1.54104 | 0.348632 |
| Chitin 1h | 76.6619 | 9.54288 | 2.02287 | 0.208134 |
| Epi 1h | 41.3698 | 5.5067 | 1.13358 | 0.1554 |
| SA 1h | 68.6002 | 14.1097 | 1.54817 | 0.301725 |
| Me-JA 1h | 118.349 | 17.3292 | 3.41256 | 0.367759 |
| Control 6h | 29.0018 | 11.9332 | 0.516997 | 0.346744 |
| ABA 6h | 259.485 | 47.9354 | 5.65059 | 0.958338 |
| ACC 6h | 15.2544 | 4.22803 | 0.315043 | 0.0854713 |
| BABA 6h | 36.2969 | 7.13404 | 0.747831 | 0.141886 |
| Chitin 6h | 22.8881 | 7.7026 | 0.449462 | 0.218885 |
| Epi 6h | 28.4281 | 8.32868 | 0.551331 | 0.225829 |
| SA 6h | 24.6575 | 7.41658 | 0.532409 | 0.288542 |
| Me-JA 6h | 35.6177 | 4.05893 | 0.856772 | 0.0983036 |
Source Transcript PGSC0003DMT400002999 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23790.1 | +1 | 6e-55 | 192 | 93/135 (69%) | Protein of unknown function (DUF607) | chr2:10125692-10127455 REVERSE LENGTH=336 |
