Probe CUST_12384_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12384_PI426222305 | JHI_St_60k_v1 | DMT400003000 | ATTCTAGATGATTCTGGAACTGTGCTTGTTCTGGGAAACGTTGTGTTCCTCAGGCCTGAT |
All Microarray Probes Designed to Gene DMG400001181
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12384_PI426222305 | JHI_St_60k_v1 | DMT400003000 | ATTCTAGATGATTCTGGAACTGTGCTTGTTCTGGGAAACGTTGTGTTCCTCAGGCCTGAT |
| CUST_12409_PI426222305 | JHI_St_60k_v1 | DMT400002999 | GTTTTGTTTACAGGTAGTGAAAACTCTACAAAGTCTATTGCCAATCCCCGCGAACCAAAA |
Microarray Signals from CUST_12384_PI426222305
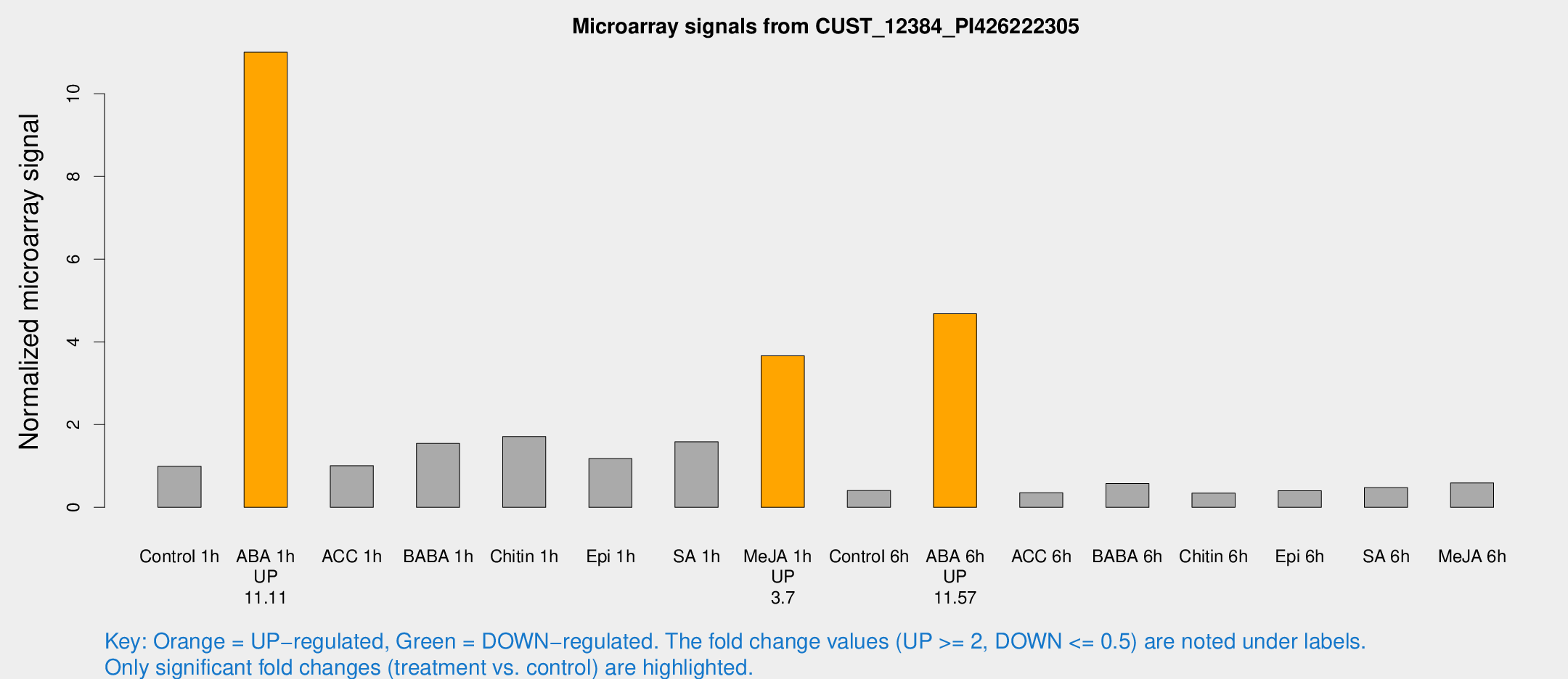
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 119.489 | 10.2367 | 0.990889 | 0.089565 |
| ABA 1h | 1215.71 | 228.004 | 11.0051 | 1.79968 |
| ACC 1h | 126.182 | 16.8219 | 1.00717 | 0.0662078 |
| BABA 1h | 184.77 | 33.2702 | 1.54501 | 0.150529 |
| Chitin 1h | 187.217 | 22.9999 | 1.7107 | 0.154525 |
| Epi 1h | 122.376 | 7.8542 | 1.1772 | 0.0756207 |
| SA 1h | 201.108 | 34.0685 | 1.58601 | 0.235365 |
| Me-JA 1h | 365.905 | 50.3391 | 3.66472 | 0.296599 |
| Control 6h | 53.8451 | 15.628 | 0.404615 | 0.107824 |
| ABA 6h | 611.719 | 94.5877 | 4.67956 | 0.551258 |
| ACC 6h | 51.1513 | 11.437 | 0.35154 | 0.0562703 |
| BABA 6h | 79.2755 | 13.4219 | 0.574727 | 0.085869 |
| Chitin 6h | 45.7486 | 8.41625 | 0.344785 | 0.0857433 |
| Epi 6h | 61.1489 | 19.5147 | 0.400567 | 0.190275 |
| SA 6h | 56.232 | 4.88628 | 0.474026 | 0.0564575 |
| Me-JA 6h | 80.5556 | 29.8078 | 0.589623 | 0.178781 |
Source Transcript PGSC0003DMT400003000 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23790.1 | +1 | 7e-104 | 317 | 181/333 (54%) | Protein of unknown function (DUF607) | chr2:10125692-10127455 REVERSE LENGTH=336 |
