Probe CUST_12184_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12184_PI426222305 | JHI_St_60k_v1 | DMT400041179 | GATCACCATTACAACTTTCATACTCCACATTATAGAGTTCACGTTTAAAGAGAGGTCAAA |
All Microarray Probes Designed to Gene DMG401015935
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12184_PI426222305 | JHI_St_60k_v1 | DMT400041179 | GATCACCATTACAACTTTCATACTCCACATTATAGAGTTCACGTTTAAAGAGAGGTCAAA |
Microarray Signals from CUST_12184_PI426222305
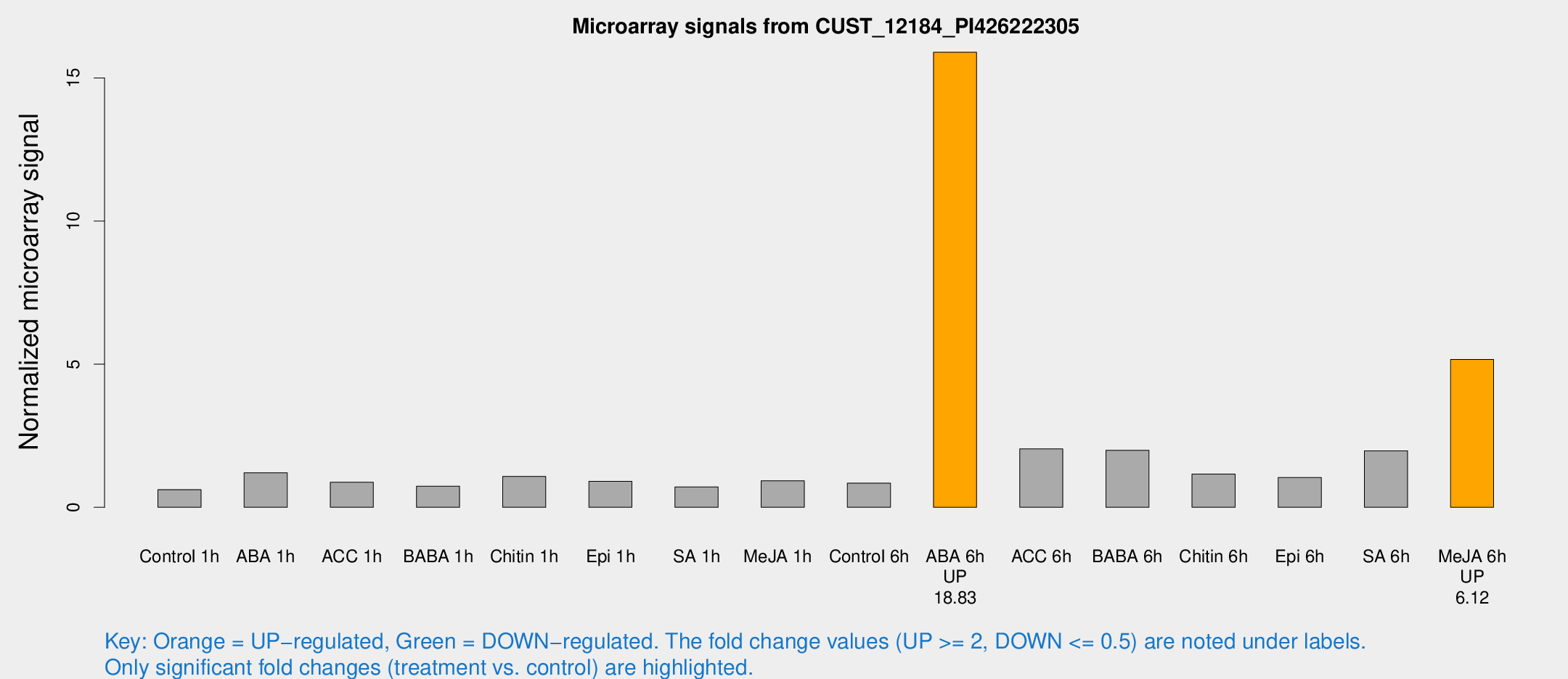
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4.98242 | 2.88849 | 0.617906 | 0.350299 |
| ABA 1h | 9.94974 | 3.62247 | 1.2084 | 0.459478 |
| ACC 1h | 7.81845 | 3.35409 | 0.873814 | 0.421145 |
| BABA 1h | 5.89581 | 3.07161 | 0.737436 | 0.390243 |
| Chitin 1h | 8.35598 | 2.92293 | 1.07979 | 0.425532 |
| Epi 1h | 6.4985 | 2.88551 | 0.90824 | 0.410743 |
| SA 1h | 6.12476 | 2.94527 | 0.712518 | 0.352757 |
| Me-JA 1h | 6.29538 | 2.99643 | 0.92735 | 0.446498 |
| Control 6h | 7.72091 | 2.99849 | 0.844184 | 0.401157 |
| ABA 6h | 147.512 | 36.4947 | 15.8979 | 2.97637 |
| ACC 6h | 19.5883 | 3.81517 | 2.04218 | 0.474781 |
| BABA 6h | 19.4405 | 4.65759 | 1.99386 | 0.405248 |
| Chitin 6h | 10.9037 | 3.37631 | 1.16021 | 0.433225 |
| Epi 6h | 10.5178 | 3.52989 | 1.0392 | 0.424419 |
| SA 6h | 21.0269 | 8.68289 | 1.97351 | 1.91046 |
| Me-JA 6h | 45.7947 | 11.1963 | 5.16571 | 1.18506 |
Source Transcript PGSC0003DMT400041179 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G33150.1 | +3 | 0.0 | 762 | 385/517 (74%) | lysine-ketoglutarate reductase/saccharopine dehydrogenase bifunctional enzyme | chr4:15985479-15991069 REVERSE LENGTH=1064 |
