Probe CUST_11860_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11860_PI426222305 | JHI_St_60k_v1 | DMT400046706 | CGTAGGAAATTACCTTTGTTTGTGGTGGCTACTCCATTGTCTAGTTAATAATTTCACTAT |
All Microarray Probes Designed to Gene DMG400018133
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11860_PI426222305 | JHI_St_60k_v1 | DMT400046706 | CGTAGGAAATTACCTTTGTTTGTGGTGGCTACTCCATTGTCTAGTTAATAATTTCACTAT |
Microarray Signals from CUST_11860_PI426222305
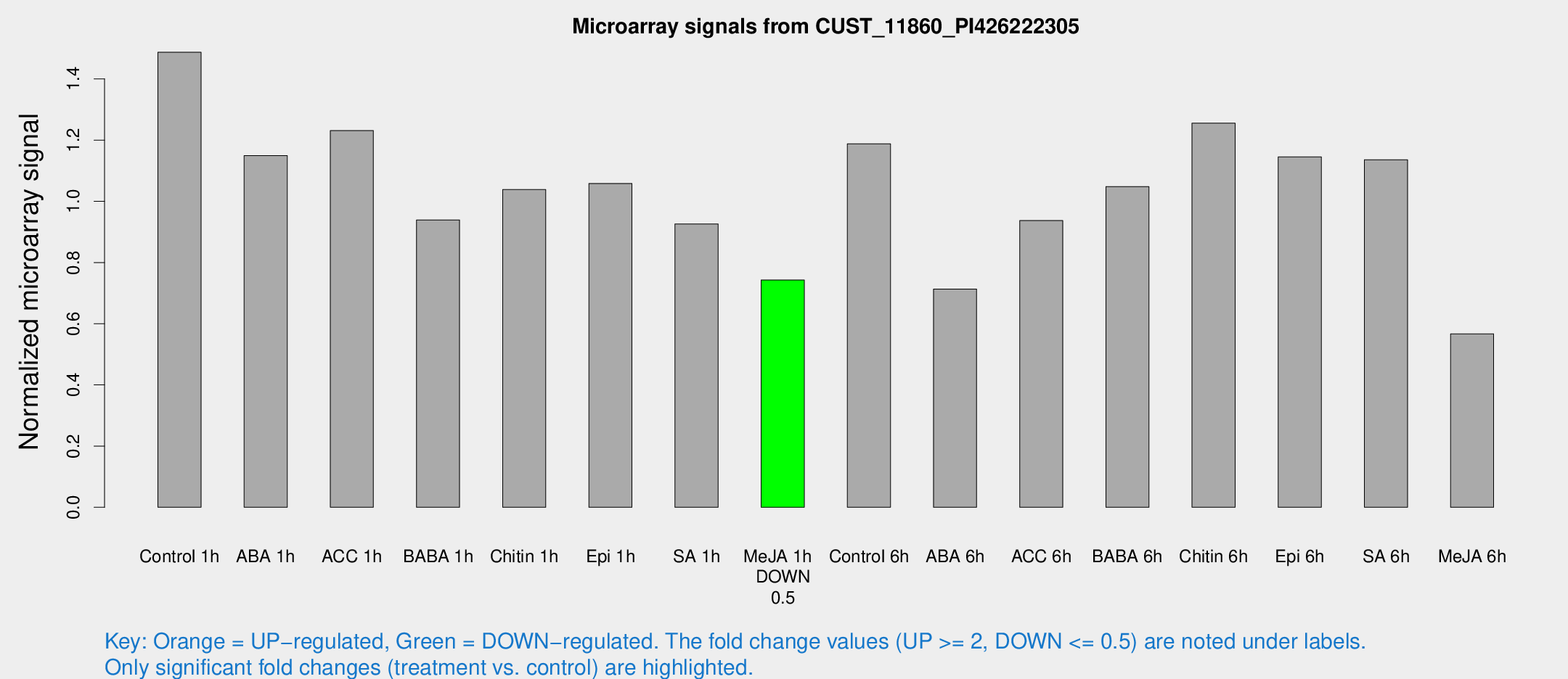
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8518.63 | 493.339 | 1.48722 | 0.0858662 |
| ABA 1h | 5852.99 | 509.476 | 1.14964 | 0.106556 |
| ACC 1h | 7277.56 | 669.169 | 1.23111 | 0.090323 |
| BABA 1h | 5409.61 | 1087.84 | 0.939062 | 0.114712 |
| Chitin 1h | 5376.42 | 494.972 | 1.03852 | 0.104572 |
| Epi 1h | 5240.89 | 303.189 | 1.05837 | 0.0611077 |
| SA 1h | 5473.2 | 479.919 | 0.9257 | 0.0534477 |
| Me-JA 1h | 3469.18 | 200.678 | 0.74281 | 0.0476717 |
| Control 6h | 7101.29 | 1498.51 | 1.18802 | 0.188335 |
| ABA 6h | 4348.95 | 281.373 | 0.71331 | 0.0411862 |
| ACC 6h | 6233.44 | 758.185 | 0.936985 | 0.0644647 |
| BABA 6h | 6831.14 | 986.532 | 1.04816 | 0.140576 |
| Chitin 6h | 7698.73 | 744.15 | 1.25526 | 0.0797067 |
| Epi 6h | 7427.87 | 642.33 | 1.14517 | 0.0910252 |
| SA 6h | 6777.5 | 1584.76 | 1.13602 | 0.168909 |
| Me-JA 6h | 3525.35 | 1041.72 | 0.566723 | 0.127654 |
Source Transcript PGSC0003DMT400046706 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G48280.1 | +1 | 4e-106 | 327 | 173/409 (42%) | cytochrome P450, family 71, subfamily A, polypeptide 25 | chr3:17879594-17881164 FORWARD LENGTH=490 |
