Probe CUST_11790_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11790_PI426222305 | JHI_St_60k_v1 | DMT400046778 | TTTACATGTAAAGAACAATCACCTTATGAAAACTCCCATCAATTCAAAGACGTACGCGTG |
All Microarray Probes Designed to Gene DMG400018163
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11790_PI426222305 | JHI_St_60k_v1 | DMT400046778 | TTTACATGTAAAGAACAATCACCTTATGAAAACTCCCATCAATTCAAAGACGTACGCGTG |
Microarray Signals from CUST_11790_PI426222305
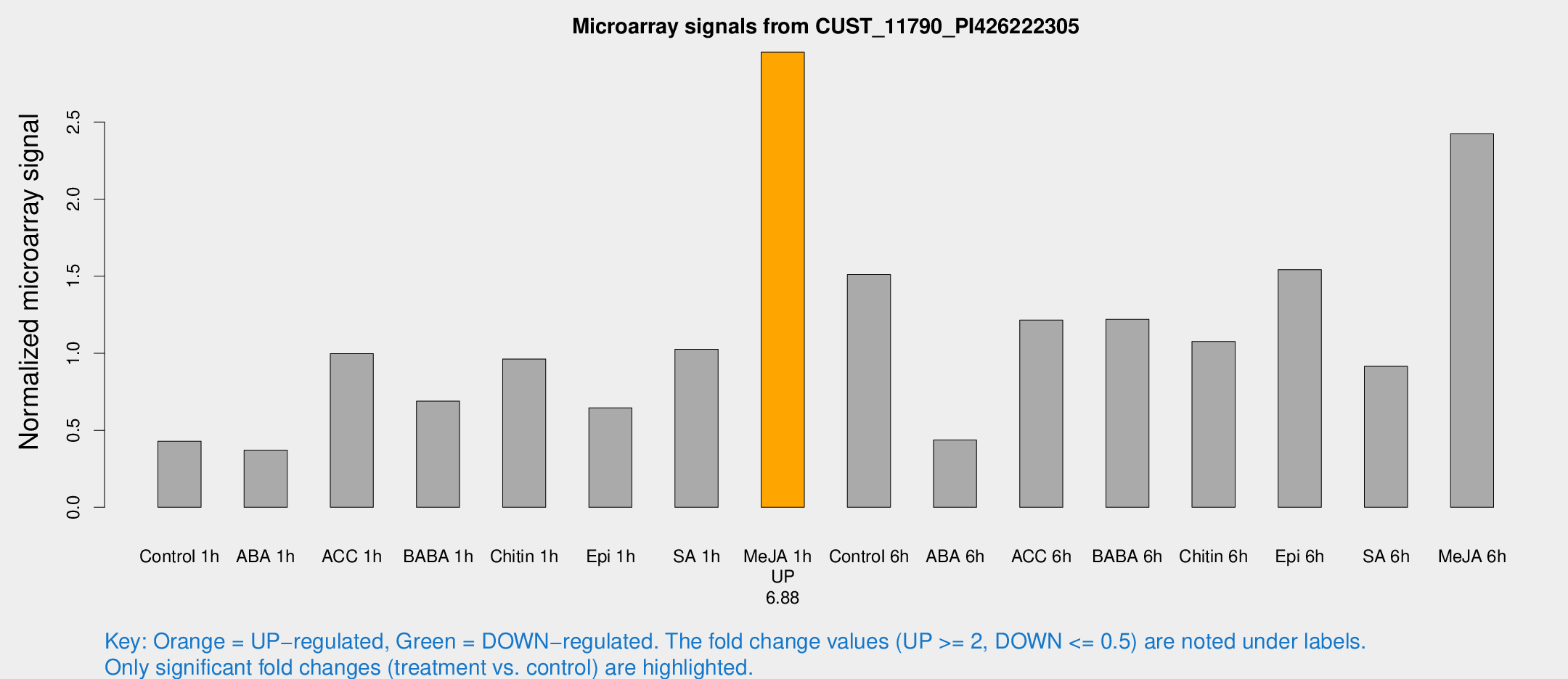
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.49829 | 3.3469 | 0.429394 | 0.211092 |
| ABA 1h | 5.48853 | 3.17861 | 0.371243 | 0.214991 |
| ACC 1h | 17.472 | 3.7021 | 0.997095 | 0.224997 |
| BABA 1h | 12.138 | 3.58956 | 0.689185 | 0.253731 |
| Chitin 1h | 15.6251 | 4.2001 | 0.962293 | 0.386408 |
| Epi 1h | 10.4433 | 3.58645 | 0.645624 | 0.265815 |
| SA 1h | 18.7495 | 4.24388 | 1.02607 | 0.280838 |
| Me-JA 1h | 42.1067 | 8.4671 | 2.9537 | 0.601712 |
| Control 6h | 25.4497 | 3.91939 | 1.51006 | 0.237987 |
| ABA 6h | 7.99848 | 3.76229 | 0.437231 | 0.219312 |
| ACC 6h | 23.4464 | 4.45119 | 1.21465 | 0.227932 |
| BABA 6h | 25.8492 | 8.20092 | 1.21989 | 0.439924 |
| Chitin 6h | 20.7794 | 5.8946 | 1.07626 | 0.313784 |
| Epi 6h | 29.1009 | 4.44606 | 1.54249 | 0.232989 |
| SA 6h | 15.2945 | 3.8958 | 0.915084 | 0.240933 |
| Me-JA 6h | 41.1163 | 5.86644 | 2.42412 | 0.539753 |
Source Transcript PGSC0003DMT400046778 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G20140.1 | +3 | 0.0 | 700 | 417/871 (48%) | Leucine-rich repeat transmembrane protein kinase | chr4:10884220-10888045 FORWARD LENGTH=1249 |
