Probe CUST_11733_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11733_PI426222305 | JHI_St_60k_v1 | DMT400046637 | TGATTGTAAACAGAGCTAGAGCTAGAGGCTAGAGCTAGTTAGTAGTATAAAGCAGTTGCT |
All Microarray Probes Designed to Gene DMG400018110
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11733_PI426222305 | JHI_St_60k_v1 | DMT400046637 | TGATTGTAAACAGAGCTAGAGCTAGAGGCTAGAGCTAGTTAGTAGTATAAAGCAGTTGCT |
Microarray Signals from CUST_11733_PI426222305
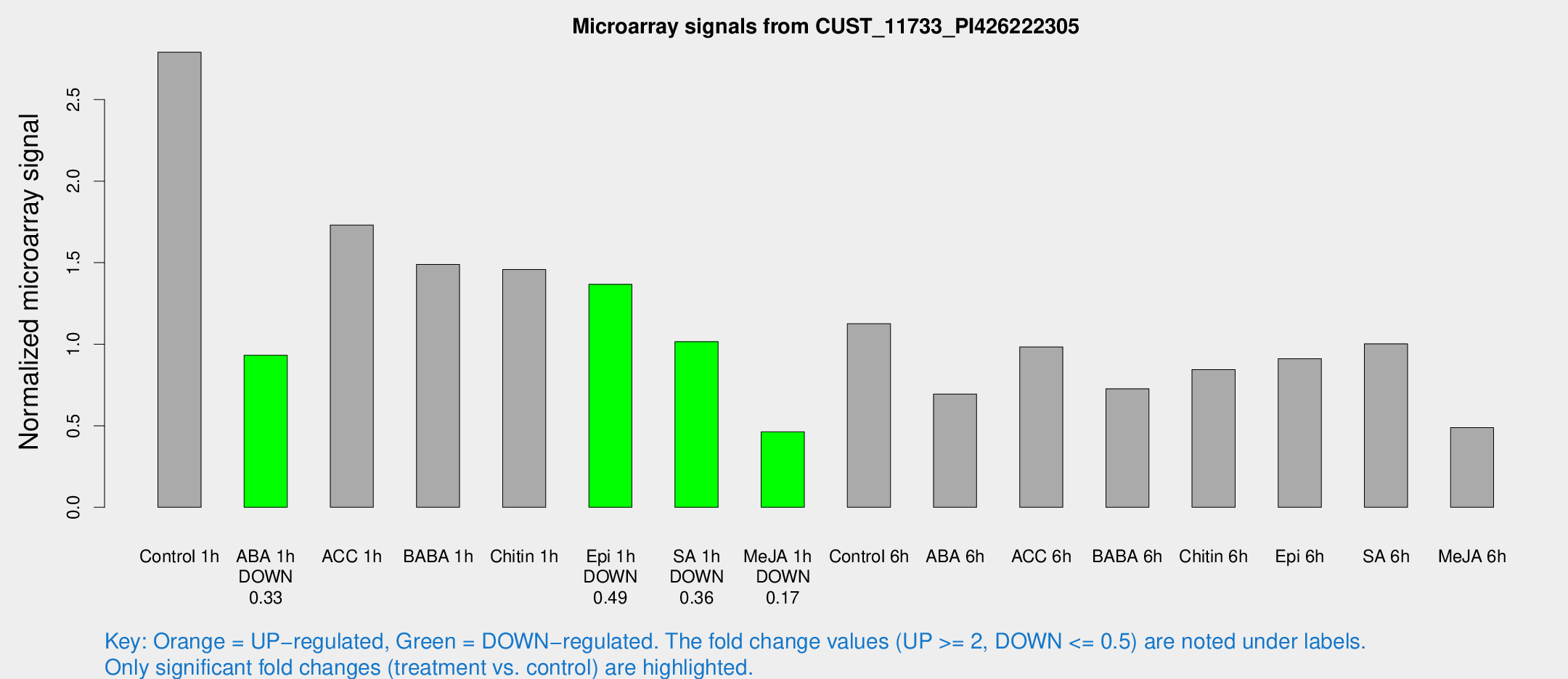
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 153.009 | 12.6791 | 2.79008 | 0.174961 |
| ABA 1h | 45.8785 | 6.81169 | 0.931585 | 0.180085 |
| ACC 1h | 102.194 | 24.8673 | 1.73079 | 0.344301 |
| BABA 1h | 80.0308 | 12.5857 | 1.48936 | 0.191662 |
| Chitin 1h | 73.418 | 11.7229 | 1.45798 | 0.195142 |
| Epi 1h | 64.59 | 5.12306 | 1.36714 | 0.108014 |
| SA 1h | 58.9937 | 11.8366 | 1.01593 | 0.203758 |
| Me-JA 1h | 22.8563 | 7.47404 | 0.462466 | 0.136462 |
| Control 6h | 66.9714 | 20.5836 | 1.12602 | 0.323634 |
| ABA 6h | 41.7786 | 7.54589 | 0.694545 | 0.186044 |
| ACC 6h | 66.1563 | 18.7805 | 0.983456 | 0.12557 |
| BABA 6h | 47.8306 | 12.4157 | 0.726854 | 0.194019 |
| Chitin 6h | 52.0491 | 13.2963 | 0.84395 | 0.209527 |
| Epi 6h | 56.3708 | 5.54239 | 0.910697 | 0.088359 |
| SA 6h | 55.8042 | 10.3059 | 1.00251 | 0.127226 |
| Me-JA 6h | 28.9787 | 8.89821 | 0.488645 | 0.108419 |
Source Transcript PGSC0003DMT400046637 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G24670.1 | +2 | 3e-145 | 432 | 213/424 (50%) | tryptophan aminotransferase related 2 | chr4:12727940-12730694 REVERSE LENGTH=440 |
