Probe CUST_11269_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11269_PI426222305 | JHI_St_60k_v1 | DMT400078342 | AATGTTCAGGGTTCTTCCCTTCACTACAACTGTGTACCCTTGTGCAATTACAGCTCCAAG |
All Microarray Probes Designed to Gene DMG400030501
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11269_PI426222305 | JHI_St_60k_v1 | DMT400078342 | AATGTTCAGGGTTCTTCCCTTCACTACAACTGTGTACCCTTGTGCAATTACAGCTCCAAG |
Microarray Signals from CUST_11269_PI426222305
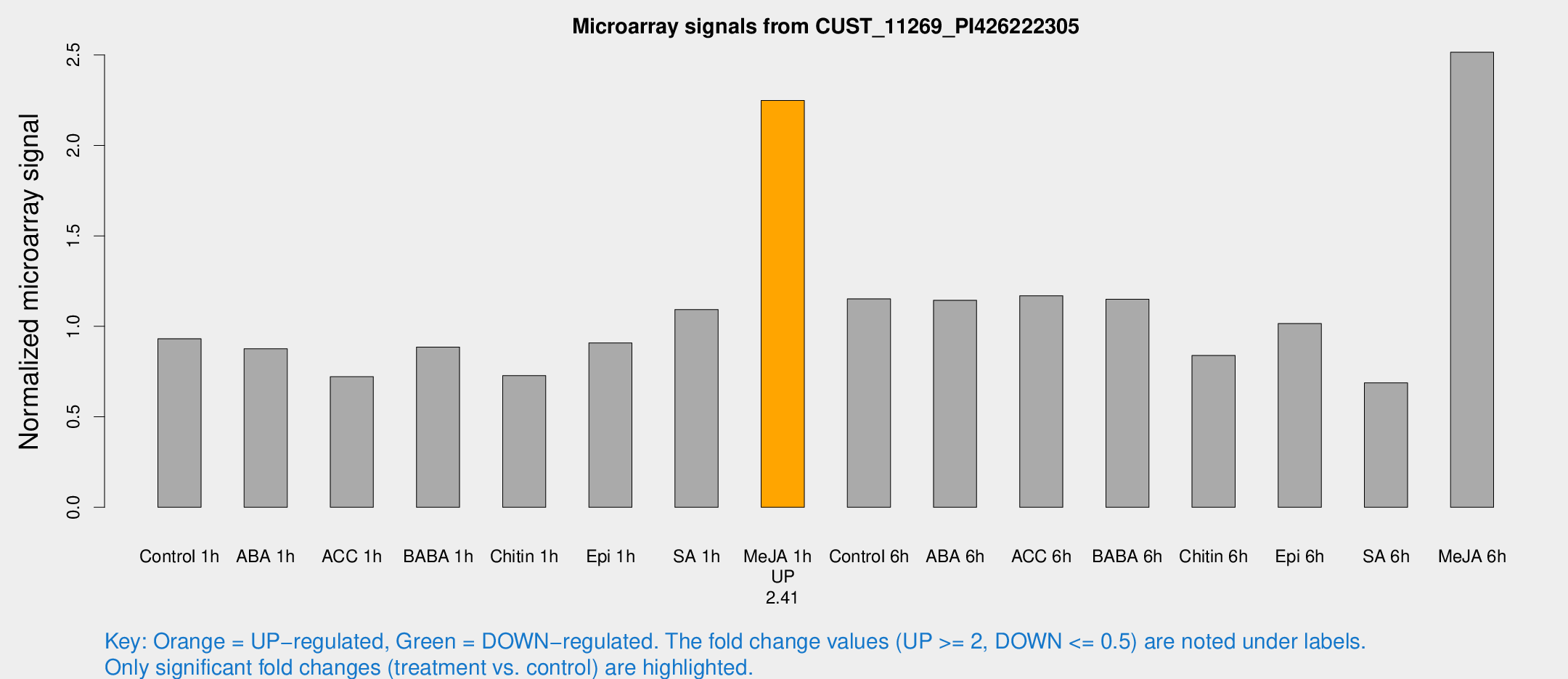
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 151.897 | 22.8913 | 0.931383 | 0.0945991 |
| ABA 1h | 123.897 | 7.81365 | 0.876013 | 0.0607596 |
| ACC 1h | 136.51 | 47.3047 | 0.72166 | 0.256558 |
| BABA 1h | 142.814 | 30.8153 | 0.885709 | 0.123759 |
| Chitin 1h | 105.644 | 10.9231 | 0.727449 | 0.0926557 |
| Epi 1h | 126.513 | 11.4397 | 0.908442 | 0.0932513 |
| SA 1h | 179.543 | 10.8679 | 1.09262 | 0.0660322 |
| Me-JA 1h | 293.879 | 17.3383 | 2.2485 | 0.15079 |
| Control 6h | 198.505 | 47.4722 | 1.15217 | 0.22933 |
| ABA 6h | 204.321 | 41.7965 | 1.14427 | 0.230102 |
| ACC 6h | 218.857 | 31.7731 | 1.16928 | 0.122491 |
| BABA 6h | 211.75 | 35.6269 | 1.14953 | 0.158928 |
| Chitin 6h | 145.241 | 18.7474 | 0.838939 | 0.118165 |
| Epi 6h | 185.781 | 23.0823 | 1.01528 | 0.19253 |
| SA 6h | 115.069 | 28.3677 | 0.688021 | 0.121193 |
| Me-JA 6h | 410.398 | 62.0053 | 2.51481 | 0.237209 |
Source Transcript PGSC0003DMT400078342 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G01350.1 | +3 | 4e-177 | 522 | 266/335 (79%) | quinolinate phoshoribosyltransferase | chr2:165332-167209 REVERSE LENGTH=348 |
