Probe CUST_10816_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10816_PI426222305 | JHI_St_60k_v1 | DMT400031803 | GGACCGGACCATCTTATTTTCGATCTCTTGATTCTTGCTGTTCATTATTTTAAATTGATC |
All Microarray Probes Designed to Gene DMG400012201
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10816_PI426222305 | JHI_St_60k_v1 | DMT400031803 | GGACCGGACCATCTTATTTTCGATCTCTTGATTCTTGCTGTTCATTATTTTAAATTGATC |
Microarray Signals from CUST_10816_PI426222305
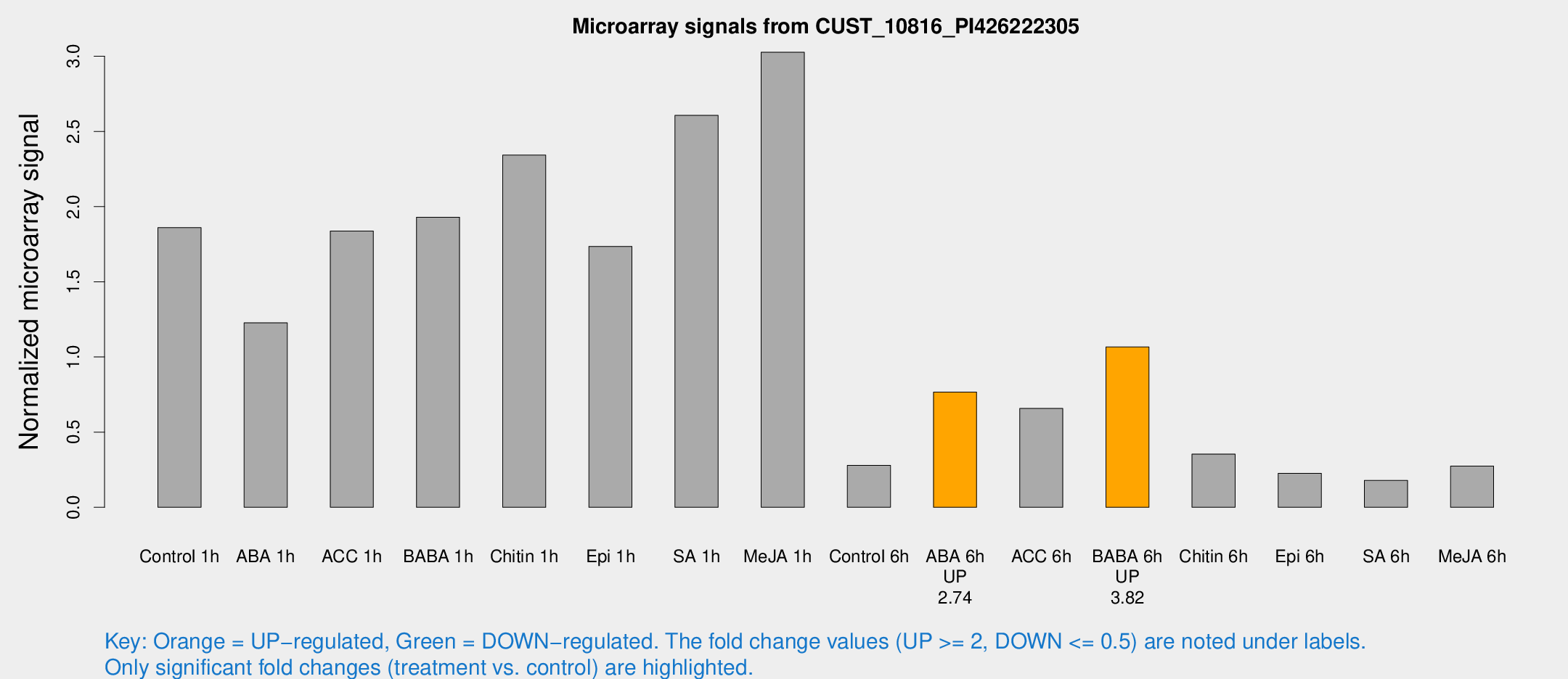
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5969.32 | 345.665 | 1.8605 | 0.10742 |
| ABA 1h | 3583.1 | 597.879 | 1.22688 | 0.22356 |
| ACC 1h | 6846.59 | 2028.06 | 1.83792 | 0.600843 |
| BABA 1h | 6200.8 | 1176.27 | 1.92968 | 0.209851 |
| Chitin 1h | 6745.1 | 389.712 | 2.34298 | 0.248873 |
| Epi 1h | 4818.61 | 278.988 | 1.7355 | 0.100205 |
| SA 1h | 8649.99 | 844.298 | 2.60729 | 0.232368 |
| Me-JA 1h | 7971.29 | 722.859 | 3.0271 | 0.174774 |
| Control 6h | 975.234 | 262.599 | 0.279405 | 0.0601076 |
| ABA 6h | 2750.02 | 603.791 | 0.766339 | 0.144328 |
| ACC 6h | 2461.88 | 362.519 | 0.657392 | 0.0379682 |
| BABA 6h | 4170.6 | 1279.3 | 1.0663 | 0.285772 |
| Chitin 6h | 1225.89 | 162.617 | 0.354052 | 0.0548112 |
| Epi 6h | 821.36 | 68.6204 | 0.226177 | 0.0225332 |
| SA 6h | 570.098 | 48.5355 | 0.178763 | 0.0103783 |
| Me-JA 6h | 937.264 | 252.731 | 0.274167 | 0.0510807 |
Source Transcript PGSC0003DMT400031803 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G41330.1 | +2 | 0.0 | 559 | 280/450 (62%) | BTB/POZ domain with WD40/YVTN repeat-like protein | chr5:16536452-16537828 REVERSE LENGTH=458 |
