Probe CUST_10487_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10487_PI426222305 | JHI_St_60k_v1 | DMT400031810 | CGACACTGAAGATGTTCTGACTCAAATCACAGATGATGTTCTTGAAGAATCAATTATGTA |
All Microarray Probes Designed to Gene DMG400012204
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10487_PI426222305 | JHI_St_60k_v1 | DMT400031810 | CGACACTGAAGATGTTCTGACTCAAATCACAGATGATGTTCTTGAAGAATCAATTATGTA |
| CUST_10759_PI426222305 | JHI_St_60k_v1 | DMT400031811 | CGACACTGAAGATGTTCTGACTCAAATCACAGATGATGTTCTTGAAGAATCAATTATGTA |
Microarray Signals from CUST_10487_PI426222305
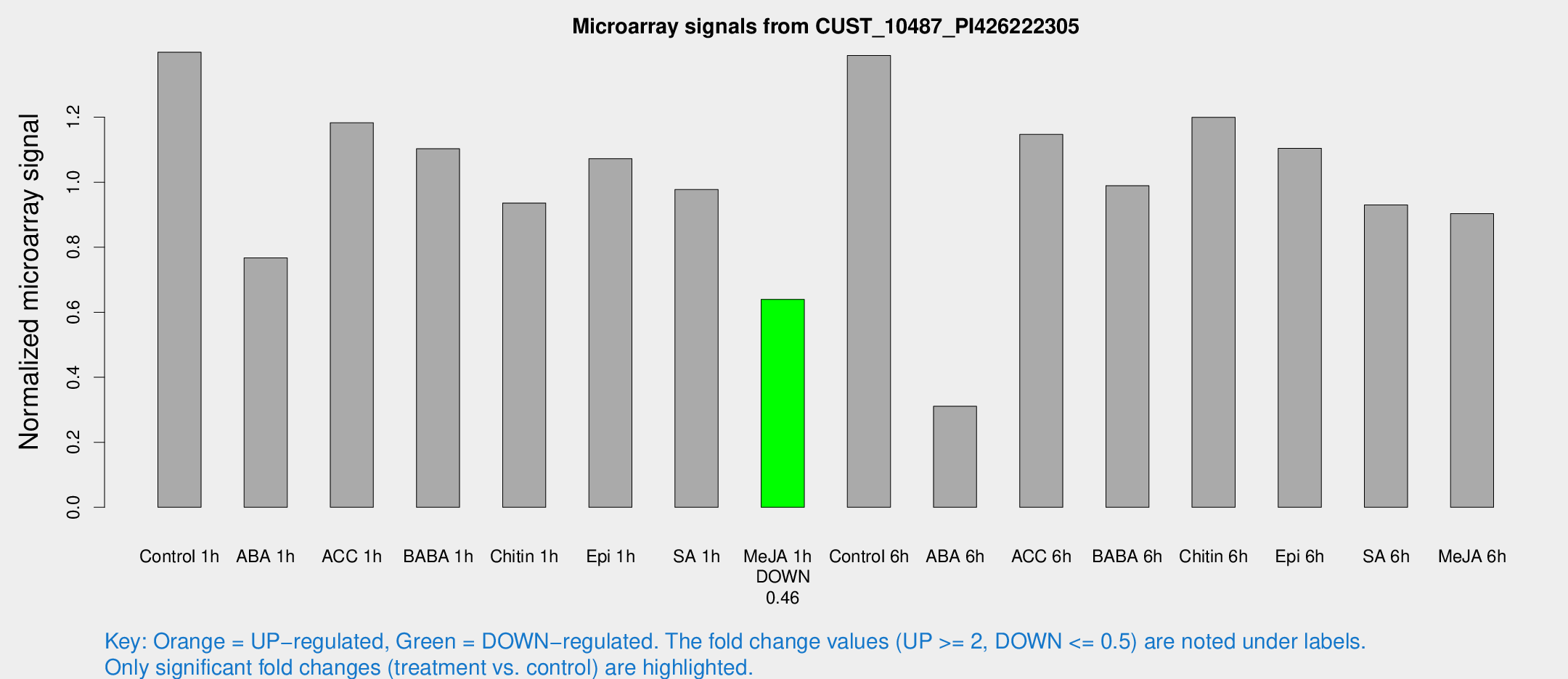
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1573.92 | 137.97 | 1.39975 | 0.0808705 |
| ABA 1h | 761.844 | 56.5873 | 0.767339 | 0.0599039 |
| ACC 1h | 1478.68 | 401.007 | 1.18255 | 0.282369 |
| BABA 1h | 1203.37 | 137.479 | 1.10275 | 0.063758 |
| Chitin 1h | 939.413 | 54.3569 | 0.935904 | 0.0654975 |
| Epi 1h | 1068.82 | 175.093 | 1.0725 | 0.183411 |
| SA 1h | 1125.45 | 74.1376 | 0.977413 | 0.06188 |
| Me-JA 1h | 583.634 | 33.9241 | 0.639672 | 0.0371314 |
| Control 6h | 1740.76 | 545.912 | 1.38949 | 0.397979 |
| ABA 6h | 382.34 | 73.8358 | 0.310614 | 0.0728408 |
| ACC 6h | 1538.9 | 332.227 | 1.14696 | 0.112265 |
| BABA 6h | 1318.84 | 353.502 | 0.989208 | 0.248629 |
| Chitin 6h | 1440.17 | 141.625 | 1.19969 | 0.09901 |
| Epi 6h | 1409.63 | 169.031 | 1.10436 | 0.181466 |
| SA 6h | 1032.91 | 77.4449 | 0.930091 | 0.053813 |
| Me-JA 6h | 1043.39 | 188.674 | 0.903181 | 0.116449 |
Source Transcript PGSC0003DMT400031810 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G00440.2 | +3 | 2e-27 | 119 | 193/884 (22%) | Protein of unknown function (DUF3741) | chr4:195176-198225 FORWARD LENGTH=835 |
