Probe CUST_9995_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9995_PI426222305 | JHI_St_60k_v1 | DMT400074982 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
All Microarray Probes Designed to Gene DMG400029165
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9995_PI426222305 | JHI_St_60k_v1 | DMT400074982 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
| CUST_9968_PI426222305 | JHI_St_60k_v1 | DMT400074981 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
| CUST_9997_PI426222305 | JHI_St_60k_v1 | DMT400074985 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
| CUST_9908_PI426222305 | JHI_St_60k_v1 | DMT400074984 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
| CUST_9990_PI426222305 | JHI_St_60k_v1 | DMT400074983 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
Microarray Signals from CUST_9995_PI426222305
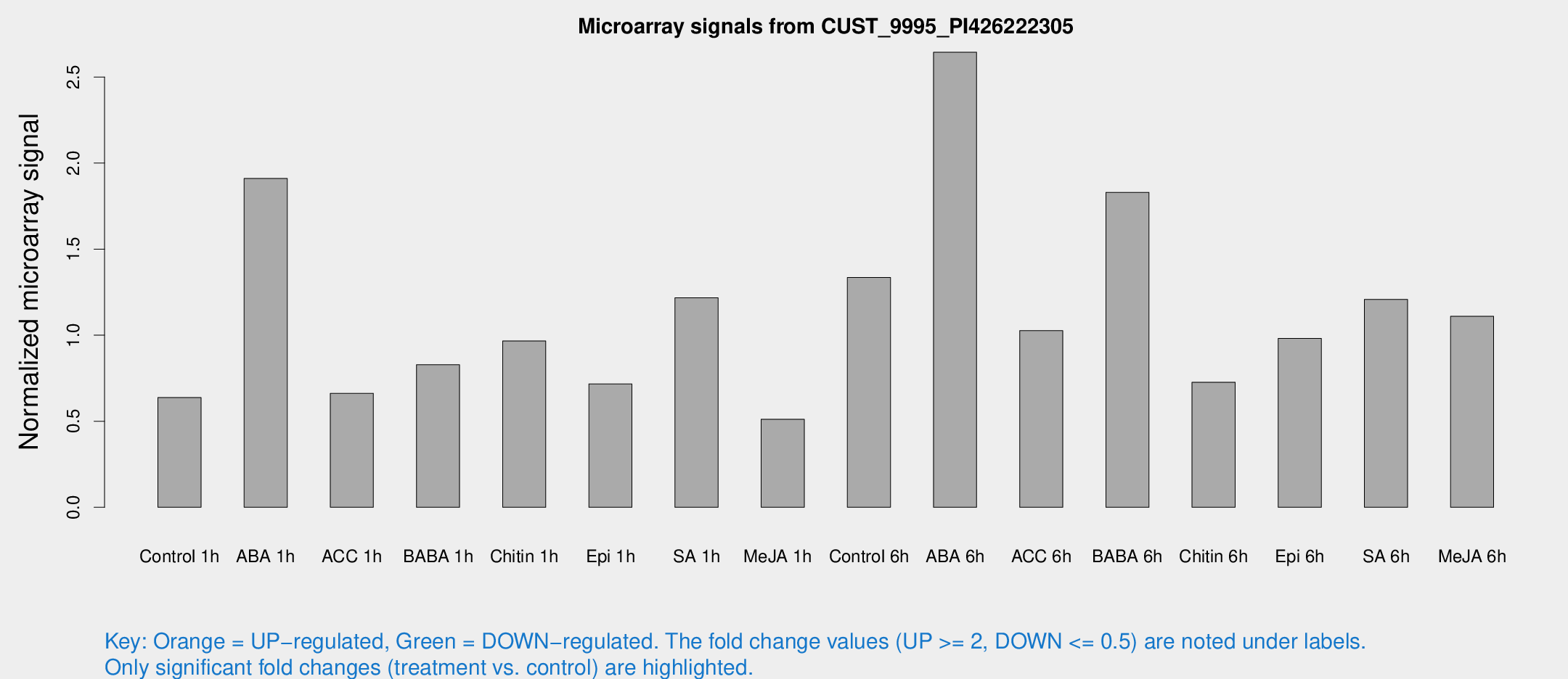
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.6666 | 3.7297 | 0.637844 | 0.219203 |
| ABA 1h | 33.661 | 4.54052 | 1.91037 | 0.236521 |
| ACC 1h | 14.5102 | 4.10907 | 0.661735 | 0.240217 |
| BABA 1h | 16.892 | 4.15278 | 0.828363 | 0.248119 |
| Chitin 1h | 18.0877 | 4.03586 | 0.96614 | 0.288888 |
| Epi 1h | 14.0996 | 5.50358 | 0.716847 | 0.281831 |
| SA 1h | 24.7452 | 3.87993 | 1.21737 | 0.197465 |
| Me-JA 1h | 9.17093 | 3.69311 | 0.511839 | 0.246583 |
| Control 6h | 29.4236 | 8.46961 | 1.33539 | 0.386917 |
| ABA 6h | 58.793 | 15.883 | 2.64412 | 0.545875 |
| ACC 6h | 25.3676 | 8.27333 | 1.02625 | 0.218276 |
| BABA 6h | 40.1129 | 4.76787 | 1.83006 | 0.217412 |
| Chitin 6h | 15.7944 | 4.34675 | 0.726789 | 0.219399 |
| Epi 6h | 22.749 | 5.29081 | 0.981032 | 0.228907 |
| SA 6h | 23.6183 | 4.29437 | 1.20826 | 0.224364 |
| Me-JA 6h | 25.2369 | 10.3809 | 1.10948 | 0.482215 |
Source Transcript PGSC0003DMT400074982 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G60580.2 | +2 | 4e-48 | 176 | 81/108 (75%) | RING/U-box superfamily protein | chr5:24354298-24356706 FORWARD LENGTH=494 |
