Probe CUST_9990_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9990_PI426222305 | JHI_St_60k_v1 | DMT400074983 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
All Microarray Probes Designed to Gene DMG400029165
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9995_PI426222305 | JHI_St_60k_v1 | DMT400074982 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
| CUST_9968_PI426222305 | JHI_St_60k_v1 | DMT400074981 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
| CUST_9997_PI426222305 | JHI_St_60k_v1 | DMT400074985 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
| CUST_9908_PI426222305 | JHI_St_60k_v1 | DMT400074984 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
| CUST_9990_PI426222305 | JHI_St_60k_v1 | DMT400074983 | TGCTTTCCTTCAAGGATCATTGCAGGAAGAAGTCTATATGAATTTGATGTGAGGTCAAAA |
Microarray Signals from CUST_9990_PI426222305
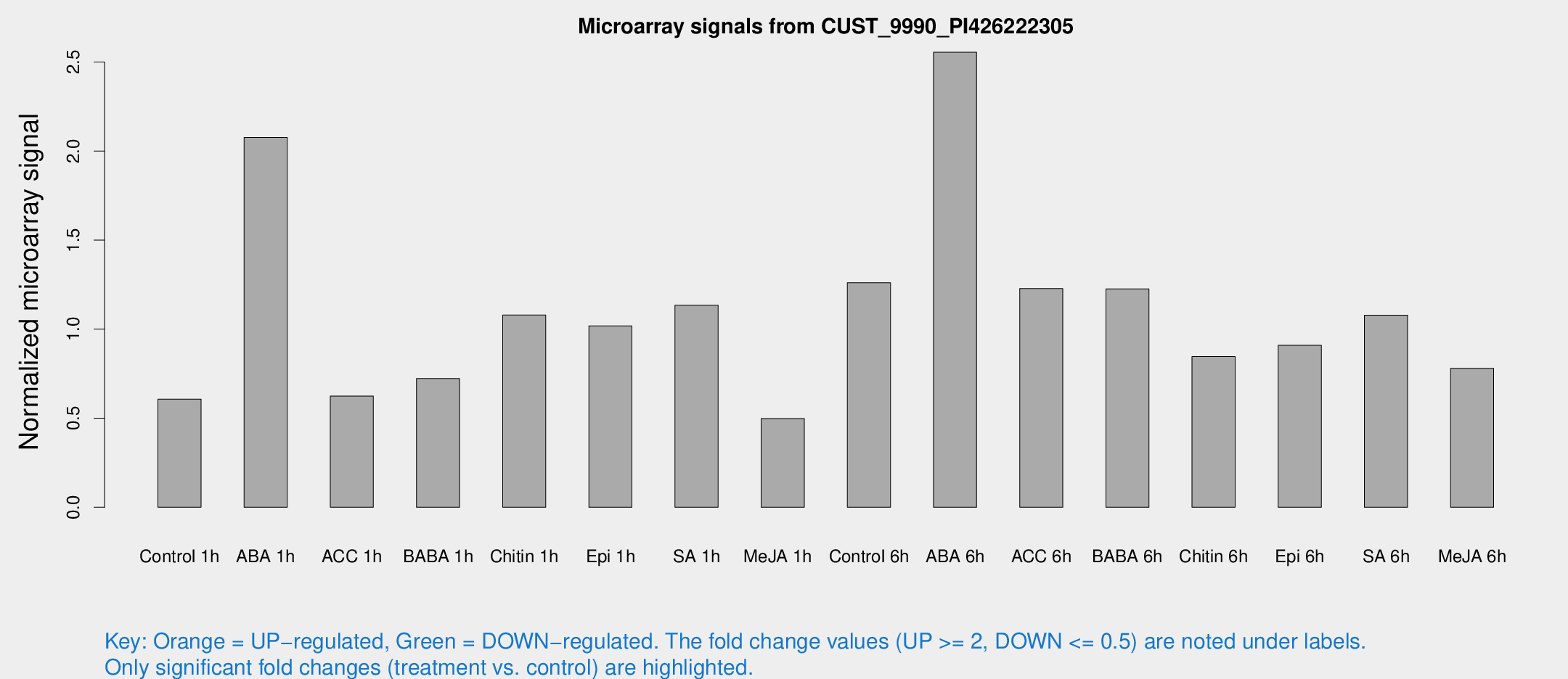
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 14.6325 | 3.65439 | 0.607303 | 0.15719 |
| ABA 1h | 47.0777 | 14.0723 | 2.07627 | 0.458693 |
| ACC 1h | 15.7957 | 4.60946 | 0.624306 | 0.218478 |
| BABA 1h | 20.5737 | 8.30758 | 0.722768 | 0.341027 |
| Chitin 1h | 23.6966 | 4.5547 | 1.07951 | 0.178237 |
| Epi 1h | 21.2283 | 3.65452 | 1.0184 | 0.182409 |
| SA 1h | 28.9172 | 5.86598 | 1.13497 | 0.296704 |
| Me-JA 1h | 10.171 | 3.73409 | 0.497714 | 0.205987 |
| Control 6h | 35.734 | 13.7025 | 1.26123 | 0.492617 |
| ABA 6h | 67.5178 | 15.5963 | 2.5547 | 0.446249 |
| ACC 6h | 34.9847 | 7.2626 | 1.22772 | 0.200258 |
| BABA 6h | 33.1681 | 4.8227 | 1.22619 | 0.175788 |
| Chitin 6h | 24.7803 | 10.0715 | 0.845971 | 0.356201 |
| Epi 6h | 27.7033 | 10.2041 | 0.909321 | 0.332289 |
| SA 6h | 28.0334 | 7.85791 | 1.07851 | 0.261889 |
| Me-JA 6h | 22.9537 | 8.3448 | 0.780241 | 0.41788 |
Source Transcript PGSC0003DMT400074983 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G60580.2 | +1 | 8e-94 | 307 | 146/210 (70%) | RING/U-box superfamily protein | chr5:24354298-24356706 FORWARD LENGTH=494 |
