Probe CUST_991_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_991_PI426222305 | JHI_St_60k_v1 | DMT400094545 | ATGAAGTGGGGGAATCTGATAGAGCAAGCAAGTTCTGCTGGTTCAGACACTACTCTCATT |
All Microarray Probes Designed to Gene DMG400044116
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_991_PI426222305 | JHI_St_60k_v1 | DMT400094545 | ATGAAGTGGGGGAATCTGATAGAGCAAGCAAGTTCTGCTGGTTCAGACACTACTCTCATT |
Microarray Signals from CUST_991_PI426222305
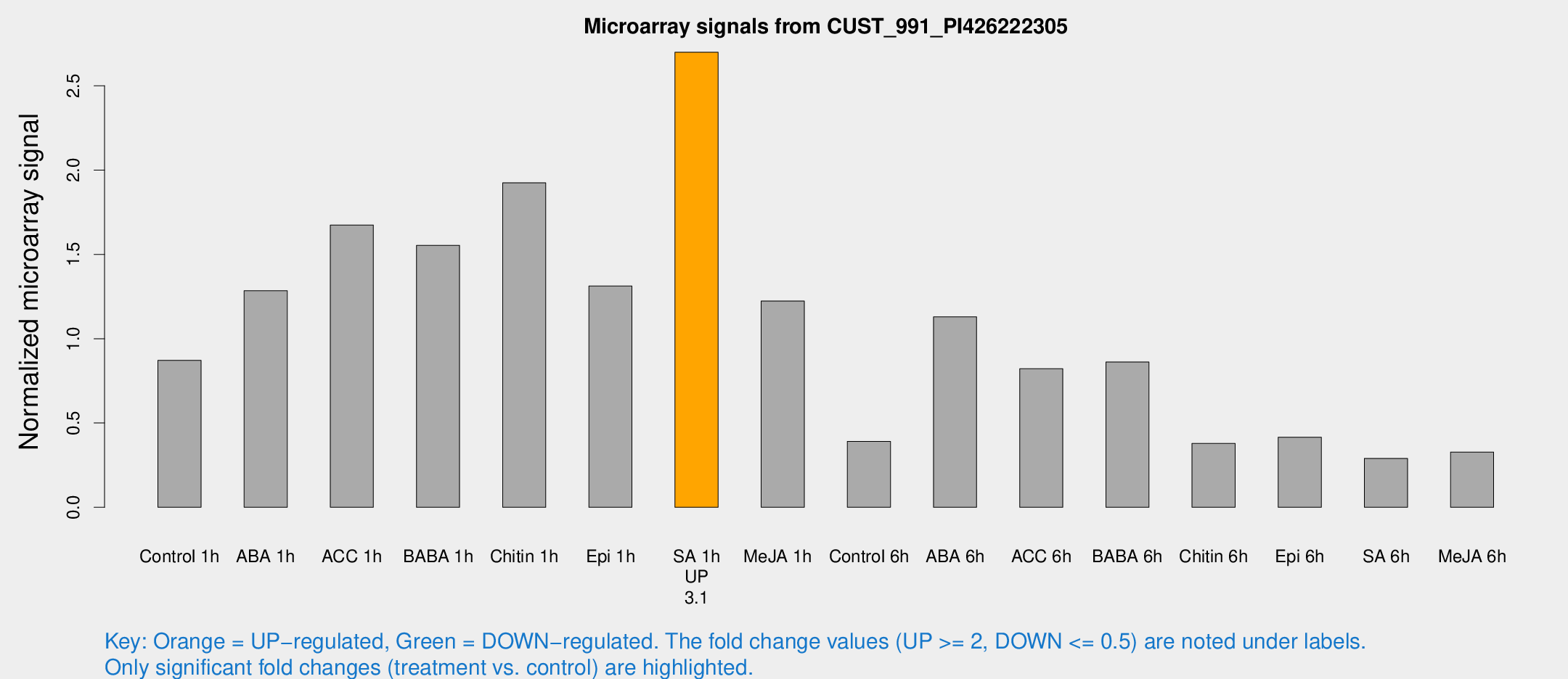
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1422.57 | 153.784 | 0.871751 | 0.0503746 |
| ABA 1h | 1867.36 | 249.568 | 1.28399 | 0.164075 |
| ACC 1h | 2816.49 | 333.182 | 1.67404 | 0.278845 |
| BABA 1h | 2461.24 | 346.143 | 1.55353 | 0.202772 |
| Chitin 1h | 2969.5 | 770.915 | 1.92427 | 0.48967 |
| Epi 1h | 1838.25 | 106.503 | 1.31218 | 0.0772068 |
| SA 1h | 4591.65 | 721.945 | 2.69875 | 0.276699 |
| Me-JA 1h | 1659.57 | 276.665 | 1.22345 | 0.301464 |
| Control 6h | 728.403 | 242.583 | 0.391226 | 0.127063 |
| ABA 6h | 1979.62 | 298.089 | 1.1292 | 0.103472 |
| ACC 6h | 1561 | 233.88 | 0.822026 | 0.13646 |
| BABA 6h | 1614.02 | 301.818 | 0.861295 | 0.167423 |
| Chitin 6h | 661.56 | 88.8354 | 0.378719 | 0.0500428 |
| Epi 6h | 781.317 | 144.858 | 0.415783 | 0.0866071 |
| SA 6h | 463.252 | 27.0346 | 0.290071 | 0.0313864 |
| Me-JA 6h | 547.995 | 112 | 0.327739 | 0.0393372 |
Source Transcript PGSC0003DMT400094545 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g089720.1 | +1 | 0.0 | 843 | 419/434 (97%) | evidence_code:10F0H1E0IEG genomic_reference:SL2.50ch02 gene_region:46036643-46038802 transcript_region:SL2.50ch02:46036643..46038802+ go_terms:GO:0016798 functional_description:Endo-beta-1 3-glucanase (AHRD V1 ***- B5THH0_PNECA); contains Interpro domain(s) IPR005200 Glycoside hydrolase, family 81 |
| TAIR PP10 | AT5G15870.1 | +1 | 0.0 | 643 | 314/448 (70%) | glycosyl hydrolase family 81 protein | chr5:5182641-5184878 REVERSE LENGTH=745 |
