Probe CUST_9540_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9540_PI426222305 | JHI_St_60k_v1 | DMT400006725 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
All Microarray Probes Designed to Gene DMG400002612
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9324_PI426222305 | JHI_St_60k_v1 | DMT400006728 | GACATACAGCTTTTGGTTTTGCCCTCATATTAGCTAACAACCTTGCTCGTTTAGCAAAAT |
| CUST_9352_PI426222305 | JHI_St_60k_v1 | DMT400006723 | CGTGTCTGGTCAGGTCTGAAATGTCAAAACATATGGCTTTACATTTTTCCTAAATACATT |
| CUST_9215_PI426222305 | JHI_St_60k_v1 | DMT400006726 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9540_PI426222305 | JHI_St_60k_v1 | DMT400006725 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9434_PI426222305 | JHI_St_60k_v1 | DMT400006724 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
Microarray Signals from CUST_9540_PI426222305
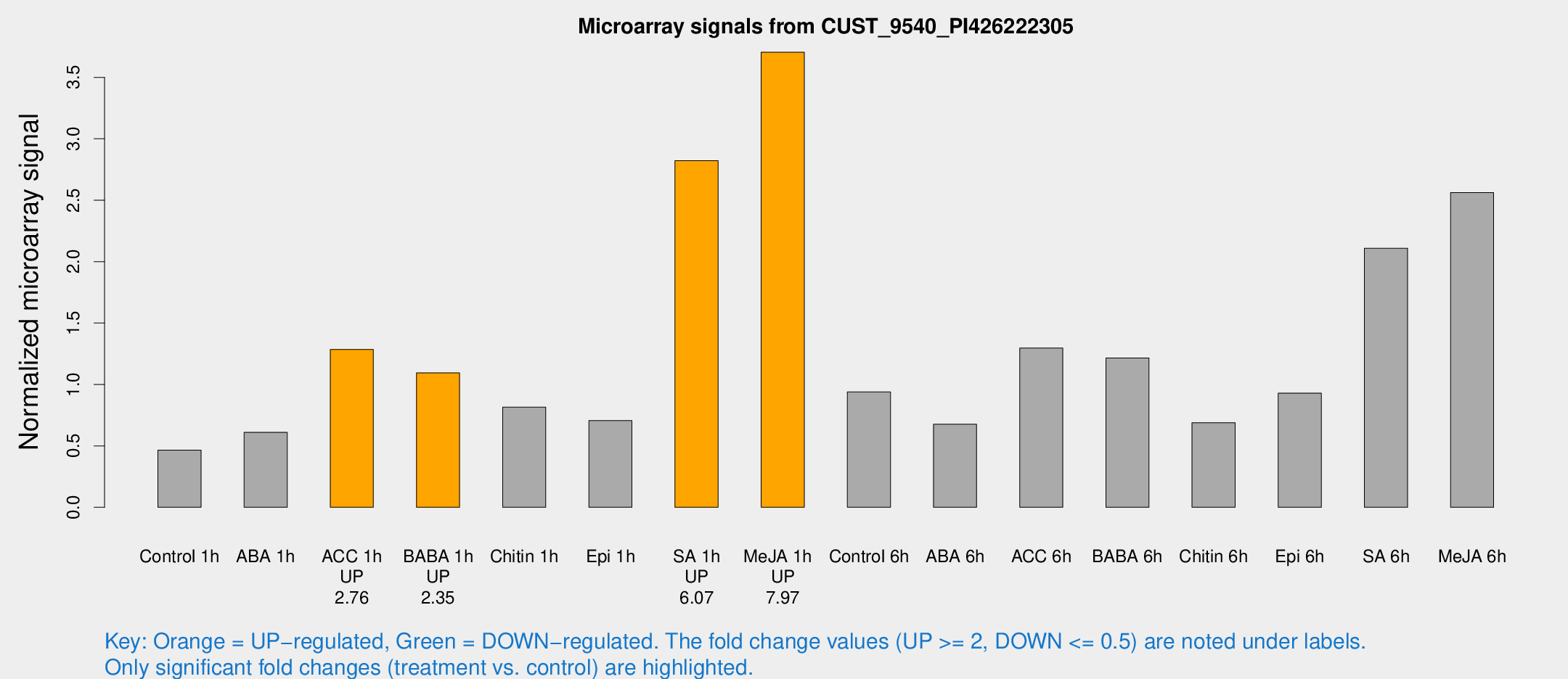
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 28.5903 | 3.74602 | 0.464937 | 0.0632221 |
| ABA 1h | 35.9346 | 10.9305 | 0.610575 | 0.150751 |
| ACC 1h | 81.2759 | 10.6544 | 1.28545 | 0.15289 |
| BABA 1h | 64.3518 | 5.30274 | 1.09448 | 0.205587 |
| Chitin 1h | 48.8384 | 15.5864 | 0.814931 | 0.254048 |
| Epi 1h | 37.3303 | 4.00593 | 0.706007 | 0.0779738 |
| SA 1h | 198.744 | 72.9735 | 2.82198 | 1.10458 |
| Me-JA 1h | 185.383 | 19.8143 | 3.70497 | 0.225153 |
| Control 6h | 75.0387 | 34.8775 | 0.938845 | 0.519105 |
| ABA 6h | 44.0324 | 4.50103 | 0.67642 | 0.0687147 |
| ACC 6h | 90.6098 | 6.84404 | 1.29619 | 0.122463 |
| BABA 6h | 83.5481 | 9.39061 | 1.21616 | 0.176616 |
| Chitin 6h | 45.7448 | 7.99696 | 0.689182 | 0.114981 |
| Epi 6h | 63.7009 | 5.45158 | 0.929309 | 0.0795117 |
| SA 6h | 130.289 | 21.2669 | 2.10827 | 0.138481 |
| Me-JA 6h | 183.925 | 77.821 | 2.5629 | 1.04636 |
Source Transcript PGSC0003DMT400006725 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79750.1 | +2 | 1e-111 | 343 | 197/251 (78%) | NADP-malic enzyme 4 | chr1:30007655-30011179 REVERSE LENGTH=646 |
