Probe CUST_9500_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9500_PI426222305 | JHI_St_60k_v1 | DMT400006584 | GCTGCCTGGAGATGTAAAAGAGTTTCTTTTGAGTGTCATAAGTTTGTGTTGATTGTACAG |
All Microarray Probes Designed to Gene DMG400002562
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9500_PI426222305 | JHI_St_60k_v1 | DMT400006584 | GCTGCCTGGAGATGTAAAAGAGTTTCTTTTGAGTGTCATAAGTTTGTGTTGATTGTACAG |
Microarray Signals from CUST_9500_PI426222305
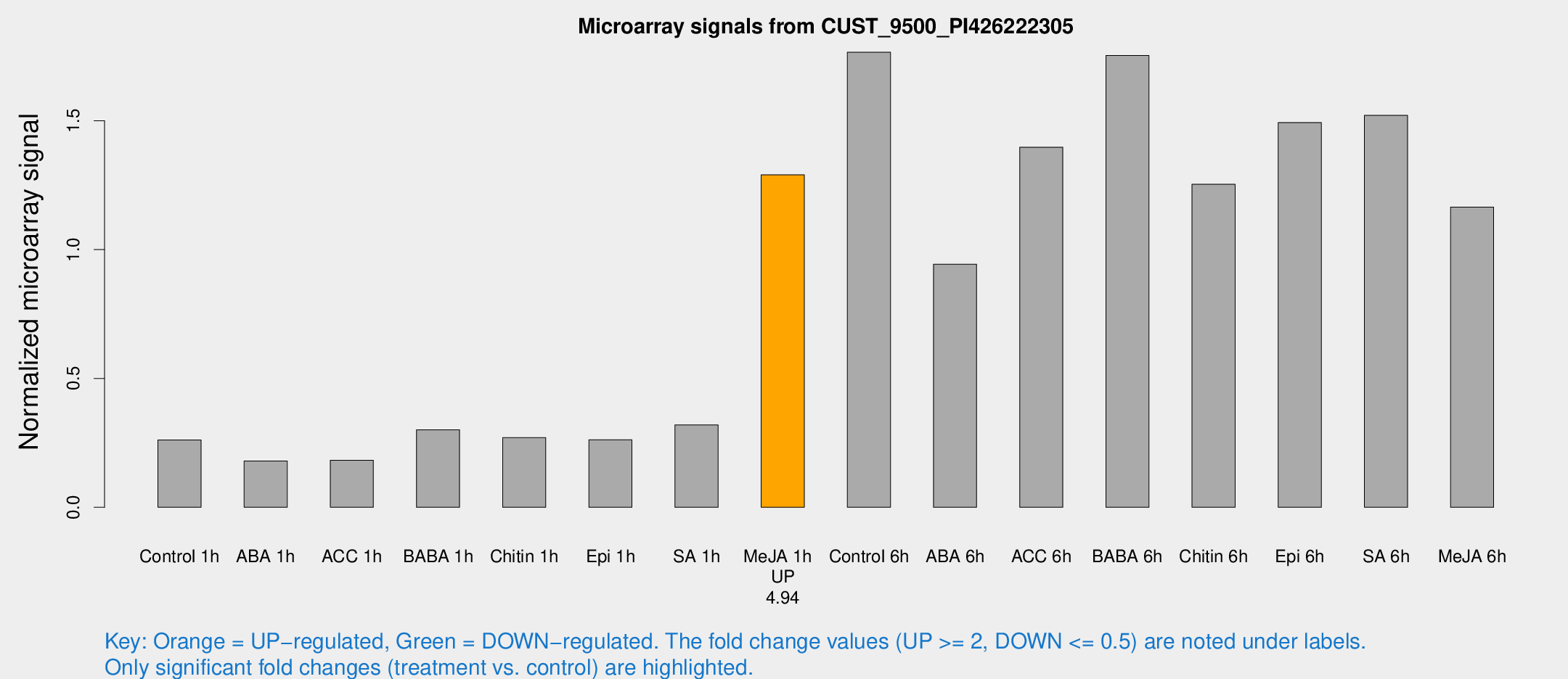
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 396.294 | 48.7966 | 0.261399 | 0.0227199 |
| ABA 1h | 263.305 | 89.3198 | 0.179491 | 0.0484045 |
| ACC 1h | 280.543 | 15.4512 | 0.182691 | 0.00995753 |
| BABA 1h | 472.753 | 123.088 | 0.300814 | 0.0646708 |
| Chitin 1h | 363.961 | 21.3831 | 0.270183 | 0.0351157 |
| Epi 1h | 359.316 | 84.3831 | 0.26227 | 0.059571 |
| SA 1h | 493.729 | 39.0243 | 0.319543 | 0.0320763 |
| Me-JA 1h | 1593.67 | 180.935 | 1.29004 | 0.118448 |
| Control 6h | 2728.81 | 460.113 | 1.76597 | 0.182237 |
| ABA 6h | 1506.27 | 103.123 | 0.943538 | 0.0822601 |
| ACC 6h | 2446.43 | 348.45 | 1.39744 | 0.0939816 |
| BABA 6h | 2949.85 | 223.629 | 1.75352 | 0.101272 |
| Chitin 6h | 2012.18 | 196.914 | 1.25337 | 0.0809998 |
| Epi 6h | 2518.79 | 145.674 | 1.49323 | 0.118223 |
| SA 6h | 2309.95 | 357.597 | 1.52081 | 0.0878522 |
| Me-JA 6h | 1835 | 394.038 | 1.16519 | 0.203144 |
Source Transcript PGSC0003DMT400006584 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G80440.1 | +2 | 1e-111 | 335 | 175/362 (48%) | Galactose oxidase/kelch repeat superfamily protein | chr1:30241763-30242827 FORWARD LENGTH=354 |
