Probe CUST_9434_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9434_PI426222305 | JHI_St_60k_v1 | DMT400006724 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
All Microarray Probes Designed to Gene DMG400002612
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9324_PI426222305 | JHI_St_60k_v1 | DMT400006728 | GACATACAGCTTTTGGTTTTGCCCTCATATTAGCTAACAACCTTGCTCGTTTAGCAAAAT |
| CUST_9352_PI426222305 | JHI_St_60k_v1 | DMT400006723 | CGTGTCTGGTCAGGTCTGAAATGTCAAAACATATGGCTTTACATTTTTCCTAAATACATT |
| CUST_9215_PI426222305 | JHI_St_60k_v1 | DMT400006726 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9540_PI426222305 | JHI_St_60k_v1 | DMT400006725 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9434_PI426222305 | JHI_St_60k_v1 | DMT400006724 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
Microarray Signals from CUST_9434_PI426222305
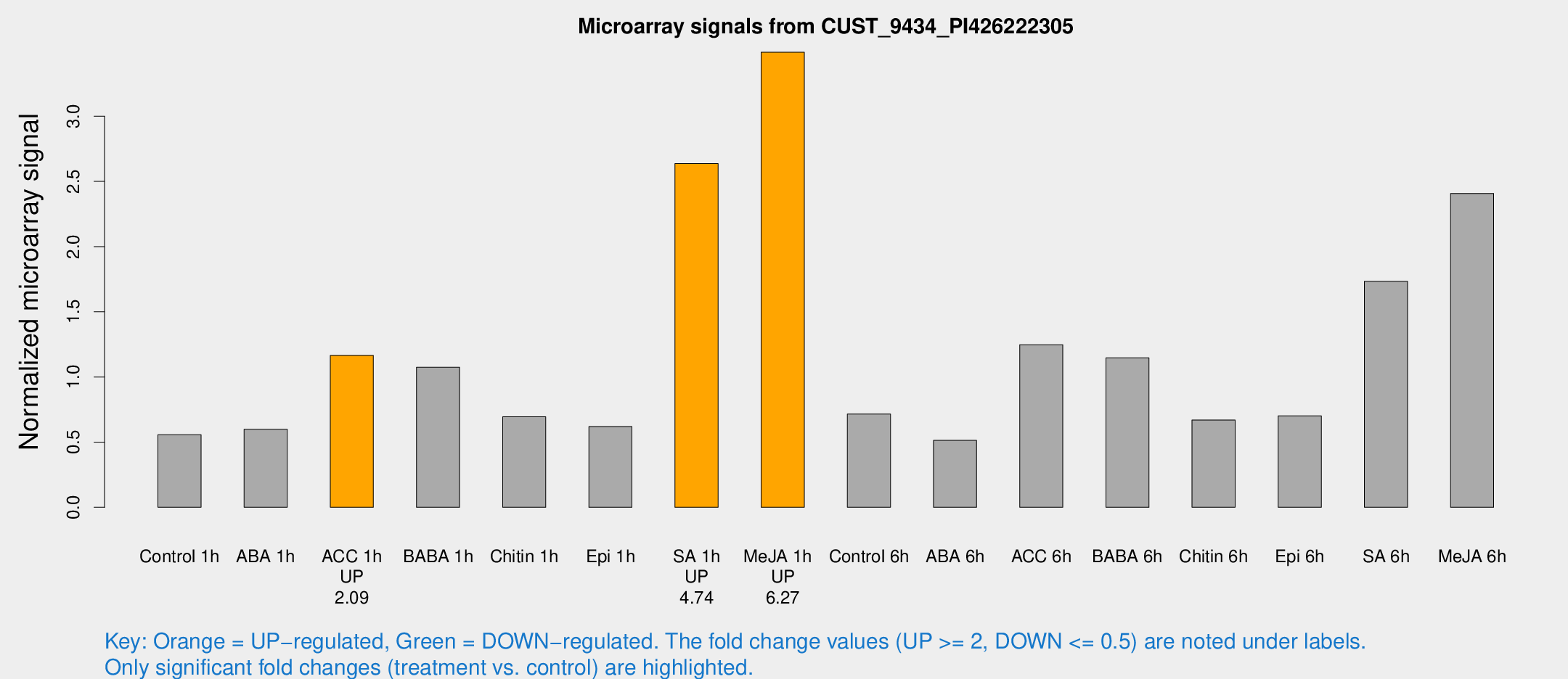
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 37.7717 | 6.87321 | 0.556388 | 0.0619091 |
| ABA 1h | 38.6406 | 12.6858 | 0.598686 | 0.172689 |
| ACC 1h | 79.3619 | 7.05336 | 1.16553 | 0.108795 |
| BABA 1h | 69.3323 | 9.28151 | 1.07443 | 0.162919 |
| Chitin 1h | 42.7685 | 8.65624 | 0.694716 | 0.131524 |
| Epi 1h | 36.0724 | 5.25744 | 0.618858 | 0.114846 |
| SA 1h | 204.907 | 80.0419 | 2.63715 | 1.03577 |
| Me-JA 1h | 189.119 | 17.6551 | 3.49095 | 0.212174 |
| Control 6h | 70.0018 | 38.8766 | 0.715517 | 0.544456 |
| ABA 6h | 36.4913 | 4.77813 | 0.51313 | 0.0590212 |
| ACC 6h | 94.7634 | 7.69125 | 1.24734 | 0.0894562 |
| BABA 6h | 84.7762 | 6.16267 | 1.14621 | 0.0856497 |
| Chitin 6h | 49.2658 | 11.0354 | 0.669541 | 0.146164 |
| Epi 6h | 54.2454 | 11.4205 | 0.702013 | 0.10496 |
| SA 6h | 117.658 | 22.3983 | 1.73333 | 0.17232 |
| Me-JA 6h | 189.165 | 81.8964 | 2.40743 | 1.00988 |
Source Transcript PGSC0003DMT400006724 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79750.1 | +2 | 9e-103 | 318 | 183/231 (79%) | NADP-malic enzyme 4 | chr1:30007655-30011179 REVERSE LENGTH=646 |
