Probe CUST_9382_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9382_PI426222305 | JHI_St_60k_v1 | DMT400006654 | GATACTGCAGTTATAACAAAAGAAGCACATTGAGACTGATTCTTGATTAGTCCATGTGTA |
All Microarray Probes Designed to Gene DMG400002590
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9170_PI426222305 | JHI_St_60k_v1 | DMT400006656 | GAATTACATTGGATCAGCCACTTGTTTATGATTGTTTTTCTGTCTTCAGTGTCAAGATTG |
| CUST_9183_PI426222305 | JHI_St_60k_v1 | DMT400006652 | GATACTGCAGTTATAACAAAAGAAGCACATTGAGACTGATTCTTGATTAGTCCATGTGTA |
| CUST_9382_PI426222305 | JHI_St_60k_v1 | DMT400006654 | GATACTGCAGTTATAACAAAAGAAGCACATTGAGACTGATTCTTGATTAGTCCATGTGTA |
Microarray Signals from CUST_9382_PI426222305
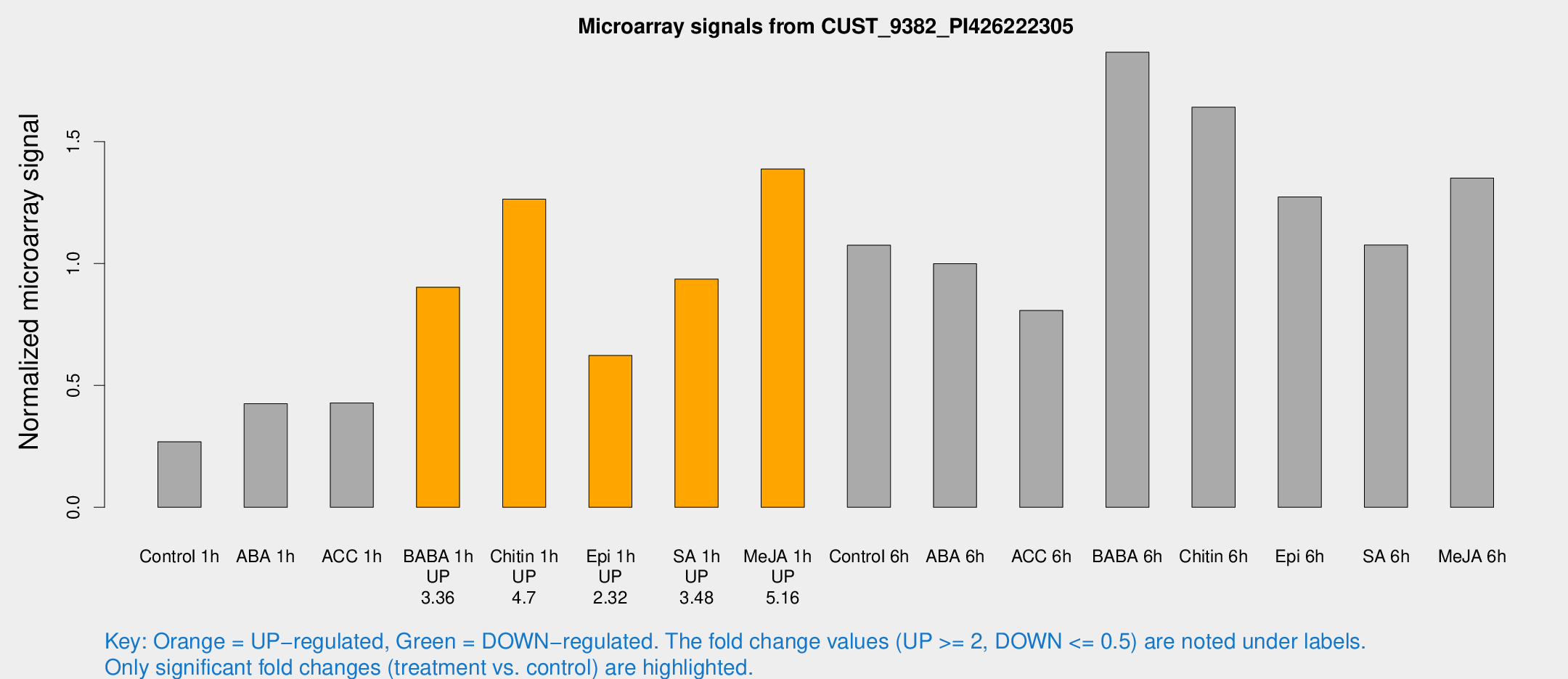
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1244.61 | 118.063 | 0.268723 | 0.0195991 |
| ABA 1h | 1825.57 | 452.124 | 0.424959 | 0.0720265 |
| ACC 1h | 2056.81 | 270.341 | 0.427894 | 0.0276189 |
| BABA 1h | 4192.06 | 841.271 | 0.902572 | 0.113732 |
| Chitin 1h | 5271.38 | 525.263 | 1.26388 | 0.211068 |
| Epi 1h | 2524.99 | 372.981 | 0.622473 | 0.0725843 |
| SA 1h | 4414.97 | 254.989 | 0.935893 | 0.0540392 |
| Me-JA 1h | 5251.38 | 539.159 | 1.38702 | 0.0954292 |
| Control 6h | 5286.93 | 1219.88 | 1.07469 | 0.198893 |
| ABA 6h | 4960.92 | 658.453 | 0.998809 | 0.076256 |
| ACC 6h | 4595.17 | 1345.36 | 0.806986 | 0.118804 |
| BABA 6h | 9902.32 | 1644.46 | 1.86634 | 0.281815 |
| Chitin 6h | 8227.22 | 1366.46 | 1.64067 | 0.211718 |
| Epi 6h | 6785.97 | 1083.58 | 1.27301 | 0.327442 |
| SA 6h | 4940.16 | 509.1 | 1.07554 | 0.0621032 |
| Me-JA 6h | 7504.56 | 2649.09 | 1.35051 | 0.6463 |
Source Transcript PGSC0003DMT400006654 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G13750.1 | +3 | 0.0 | 1367 | 644/824 (78%) | beta galactosidase 1 | chr3:4511192-4515756 FORWARD LENGTH=847 |
