Probe CUST_9352_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9352_PI426222305 | JHI_St_60k_v1 | DMT400006723 | CGTGTCTGGTCAGGTCTGAAATGTCAAAACATATGGCTTTACATTTTTCCTAAATACATT |
All Microarray Probes Designed to Gene DMG400002612
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9324_PI426222305 | JHI_St_60k_v1 | DMT400006728 | GACATACAGCTTTTGGTTTTGCCCTCATATTAGCTAACAACCTTGCTCGTTTAGCAAAAT |
| CUST_9352_PI426222305 | JHI_St_60k_v1 | DMT400006723 | CGTGTCTGGTCAGGTCTGAAATGTCAAAACATATGGCTTTACATTTTTCCTAAATACATT |
| CUST_9215_PI426222305 | JHI_St_60k_v1 | DMT400006726 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9540_PI426222305 | JHI_St_60k_v1 | DMT400006725 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9434_PI426222305 | JHI_St_60k_v1 | DMT400006724 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
Microarray Signals from CUST_9352_PI426222305
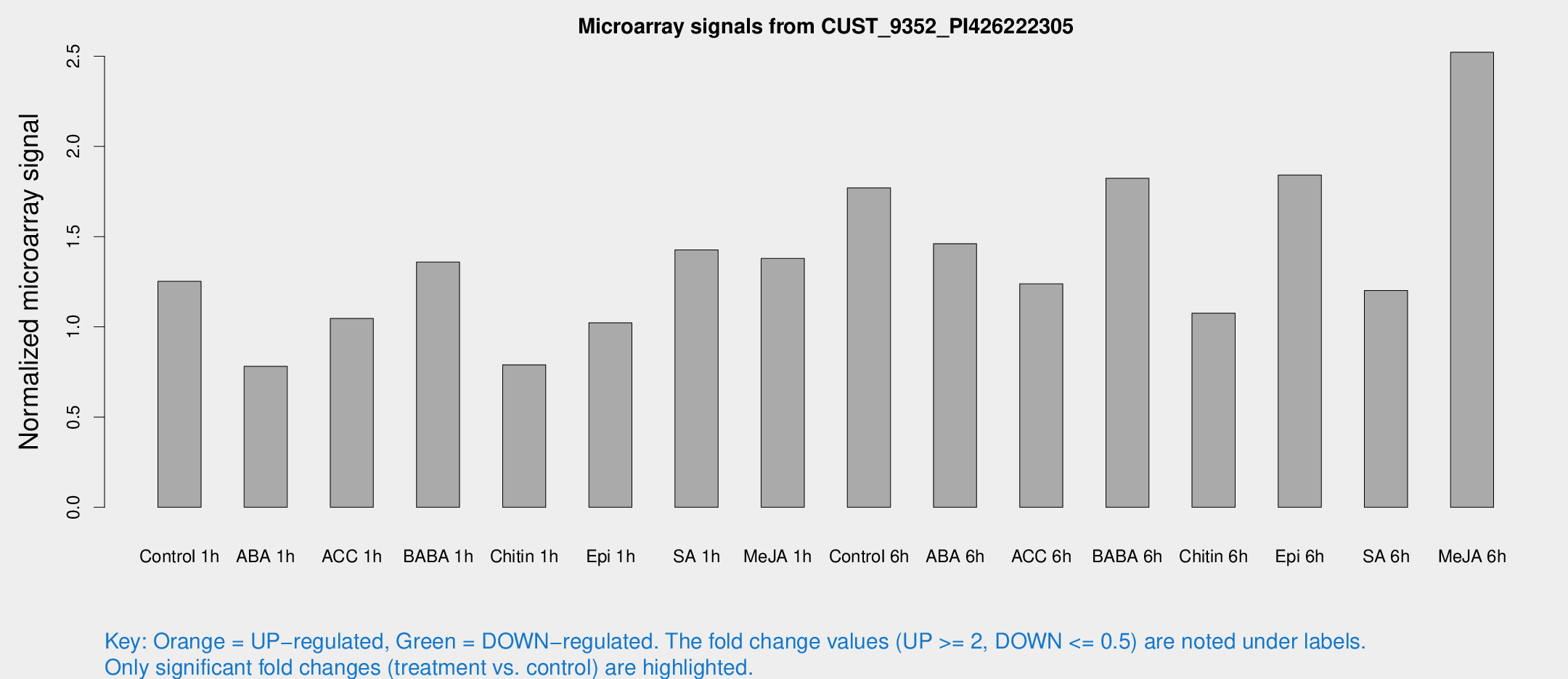
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 10.8913 | 3.43122 | 1.25221 | 0.433441 |
| ABA 1h | 5.72366 | 3.28202 | 0.781047 | 0.44797 |
| ACC 1h | 9.96811 | 3.72791 | 1.04617 | 0.49257 |
| BABA 1h | 12.6641 | 5.01086 | 1.35905 | 0.545399 |
| Chitin 1h | 5.91889 | 3.45786 | 0.789561 | 0.457785 |
| Epi 1h | 7.63966 | 3.34062 | 1.02238 | 0.496171 |
| SA 1h | 13.3284 | 4.27159 | 1.42632 | 0.566179 |
| Me-JA 1h | 12.0144 | 6.26915 | 1.37903 | 0.642588 |
| Control 6h | 17.9235 | 7.62445 | 1.76995 | 0.769081 |
| ABA 6h | 14.9671 | 5.52192 | 1.46088 | 0.55435 |
| ACC 6h | 14.7889 | 7.17088 | 1.23839 | 0.758203 |
| BABA 6h | 18.9875 | 5.48528 | 1.82387 | 0.657509 |
| Chitin 6h | 10.703 | 4.084 | 1.07571 | 0.510555 |
| Epi 6h | 23.539 | 13.4149 | 1.84173 | 1.29916 |
| SA 6h | 11.6502 | 5.03011 | 1.2013 | 0.556293 |
| Me-JA 6h | 22.3966 | 6.33726 | 2.52183 | 0.586242 |
Source Transcript PGSC0003DMT400006723 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79750.1 | +1 | 0.0 | 887 | 479/651 (74%) | NADP-malic enzyme 4 | chr1:30007655-30011179 REVERSE LENGTH=646 |
