Probe CUST_9324_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9324_PI426222305 | JHI_St_60k_v1 | DMT400006728 | GACATACAGCTTTTGGTTTTGCCCTCATATTAGCTAACAACCTTGCTCGTTTAGCAAAAT |
All Microarray Probes Designed to Gene DMG400002612
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9324_PI426222305 | JHI_St_60k_v1 | DMT400006728 | GACATACAGCTTTTGGTTTTGCCCTCATATTAGCTAACAACCTTGCTCGTTTAGCAAAAT |
| CUST_9352_PI426222305 | JHI_St_60k_v1 | DMT400006723 | CGTGTCTGGTCAGGTCTGAAATGTCAAAACATATGGCTTTACATTTTTCCTAAATACATT |
| CUST_9215_PI426222305 | JHI_St_60k_v1 | DMT400006726 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9540_PI426222305 | JHI_St_60k_v1 | DMT400006725 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9434_PI426222305 | JHI_St_60k_v1 | DMT400006724 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
Microarray Signals from CUST_9324_PI426222305
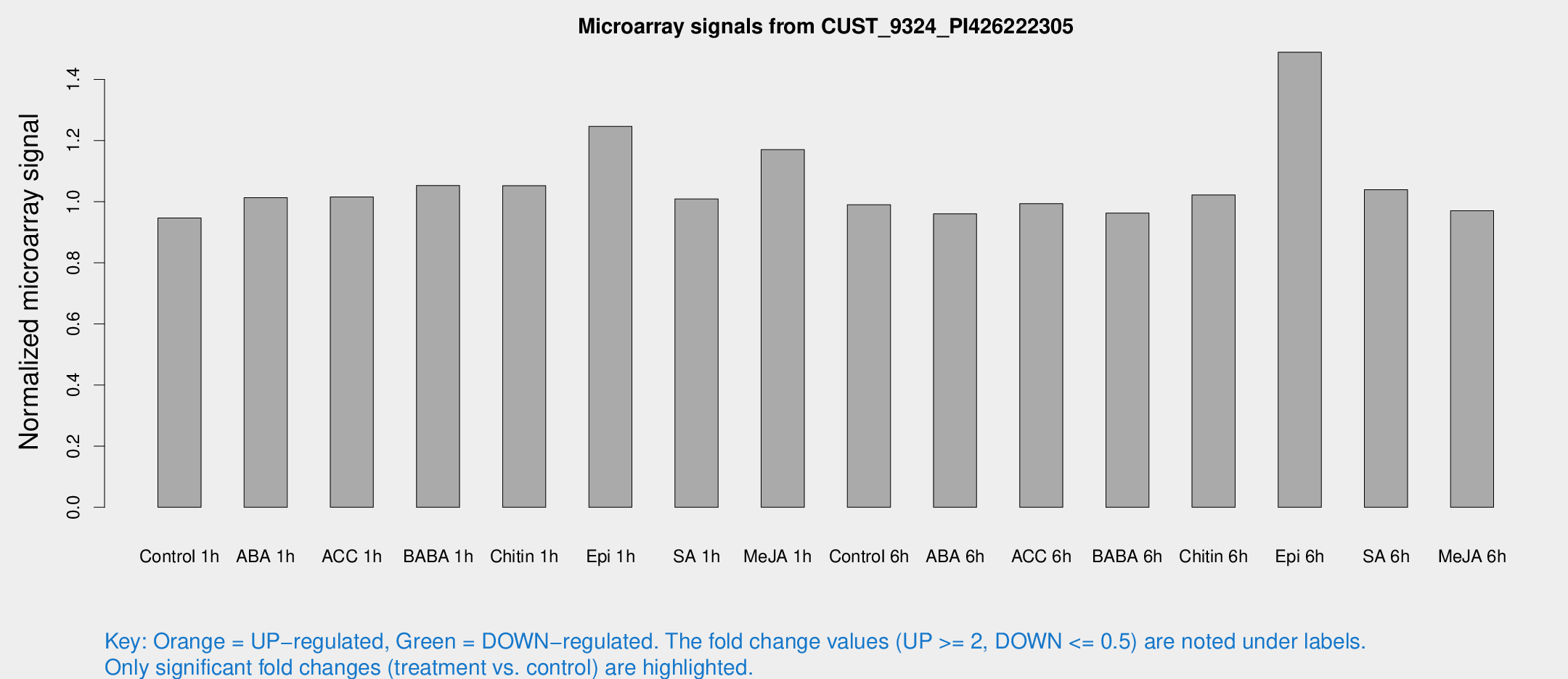
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.5143 | 3.19677 | 0.946585 | 0.548155 |
| ABA 1h | 5.21589 | 3.02322 | 1.01318 | 0.587203 |
| ACC 1h | 6.07788 | 3.52492 | 1.01558 | 0.588111 |
| BABA 1h | 5.9143 | 3.43334 | 1.05303 | 0.610402 |
| Chitin 1h | 5.46707 | 3.18154 | 1.05217 | 0.603359 |
| Epi 1h | 6.45332 | 3.1637 | 1.24655 | 0.637845 |
| SA 1h | 6.09539 | 3.15714 | 1.00877 | 0.533535 |
| Me-JA 1h | 5.58927 | 3.25705 | 1.17032 | 0.678325 |
| Control 6h | 5.77657 | 3.35222 | 0.989793 | 0.574096 |
| ABA 6h | 5.94696 | 3.44658 | 0.960451 | 0.556158 |
| ACC 6h | 6.78436 | 4.02335 | 0.993578 | 0.575351 |
| BABA 6h | 6.28123 | 3.64533 | 0.962588 | 0.5576 |
| Chitin 6h | 6.33575 | 3.67146 | 1.02222 | 0.591999 |
| Epi 6h | 13.0333 | 7.19376 | 1.48904 | 0.796062 |
| SA 6h | 5.98369 | 3.46549 | 1.03944 | 0.601918 |
| Me-JA 6h | 5.62654 | 3.26225 | 0.970633 | 0.562533 |
Source Transcript PGSC0003DMT400006728 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79750.1 | +1 | 0.0 | 563 | 282/373 (76%) | NADP-malic enzyme 4 | chr1:30007655-30011179 REVERSE LENGTH=646 |
