Probe CUST_9215_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9215_PI426222305 | JHI_St_60k_v1 | DMT400006726 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
All Microarray Probes Designed to Gene DMG400002612
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9324_PI426222305 | JHI_St_60k_v1 | DMT400006728 | GACATACAGCTTTTGGTTTTGCCCTCATATTAGCTAACAACCTTGCTCGTTTAGCAAAAT |
| CUST_9352_PI426222305 | JHI_St_60k_v1 | DMT400006723 | CGTGTCTGGTCAGGTCTGAAATGTCAAAACATATGGCTTTACATTTTTCCTAAATACATT |
| CUST_9215_PI426222305 | JHI_St_60k_v1 | DMT400006726 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9540_PI426222305 | JHI_St_60k_v1 | DMT400006725 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
| CUST_9434_PI426222305 | JHI_St_60k_v1 | DMT400006724 | TTTGTATTGGCCACTTAACATCTTATGTGTAATTCTGACCCATTGGGTCAGCACGGGCCA |
Microarray Signals from CUST_9215_PI426222305
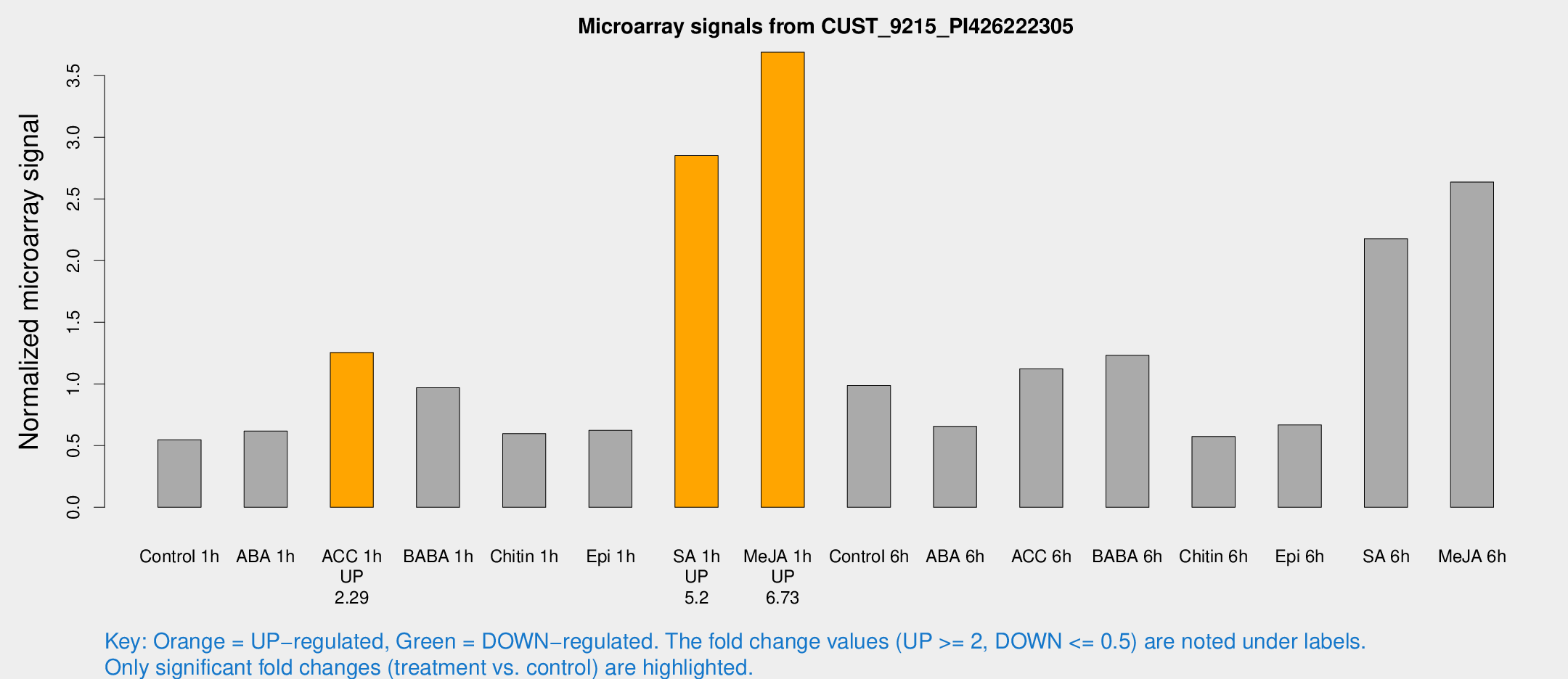
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 35.0132 | 3.94704 | 0.547968 | 0.063259 |
| ABA 1h | 38.8669 | 13.2147 | 0.617558 | 0.177542 |
| ACC 1h | 81.7131 | 6.03091 | 1.25507 | 0.092897 |
| BABA 1h | 59.3721 | 5.0312 | 0.968629 | 0.0824026 |
| Chitin 1h | 38.6629 | 12.9326 | 0.597163 | 0.215103 |
| Epi 1h | 34.7702 | 4.74901 | 0.623213 | 0.0913153 |
| SA 1h | 210.961 | 77.5377 | 2.85079 | 1.05028 |
| Me-JA 1h | 193.307 | 21.9867 | 3.68923 | 0.223929 |
| Control 6h | 81.7092 | 37.9004 | 0.987311 | 0.5253 |
| ABA 6h | 44.7276 | 4.89036 | 0.656779 | 0.0689354 |
| ACC 6h | 82.4242 | 8.3021 | 1.12248 | 0.144861 |
| BABA 6h | 87.8879 | 6.85164 | 1.23207 | 0.145926 |
| Chitin 6h | 41.2167 | 10.1883 | 0.573446 | 0.167857 |
| Epi 6h | 50.053 | 11.0463 | 0.667781 | 0.098667 |
| SA 6h | 141.299 | 24.4803 | 2.17836 | 0.162412 |
| Me-JA 6h | 193.016 | 77.9587 | 2.63683 | 0.906559 |
Source Transcript PGSC0003DMT400006726 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79750.1 | +1 | 0.0 | 902 | 479/623 (77%) | NADP-malic enzyme 4 | chr1:30007655-30011179 REVERSE LENGTH=646 |
