Probe CUST_919_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_919_PI426222305 | JHI_St_60k_v1 | DMT400025952 | TTAGCCAACTGCATTAGTTGTGTATGCAGGGGGGCAATGTTATCCAATTCACAGCAAAGC |
All Microarray Probes Designed to Gene DMG400010021
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_919_PI426222305 | JHI_St_60k_v1 | DMT400025952 | TTAGCCAACTGCATTAGTTGTGTATGCAGGGGGGCAATGTTATCCAATTCACAGCAAAGC |
| CUST_1389_PI426222305 | JHI_St_60k_v1 | DMT400025953 | TTGGTTTGGGATTGTAACAATTGAATTTGAAGCAATGTGAACAGGTGAATAAGATCTTCG |
Microarray Signals from CUST_919_PI426222305
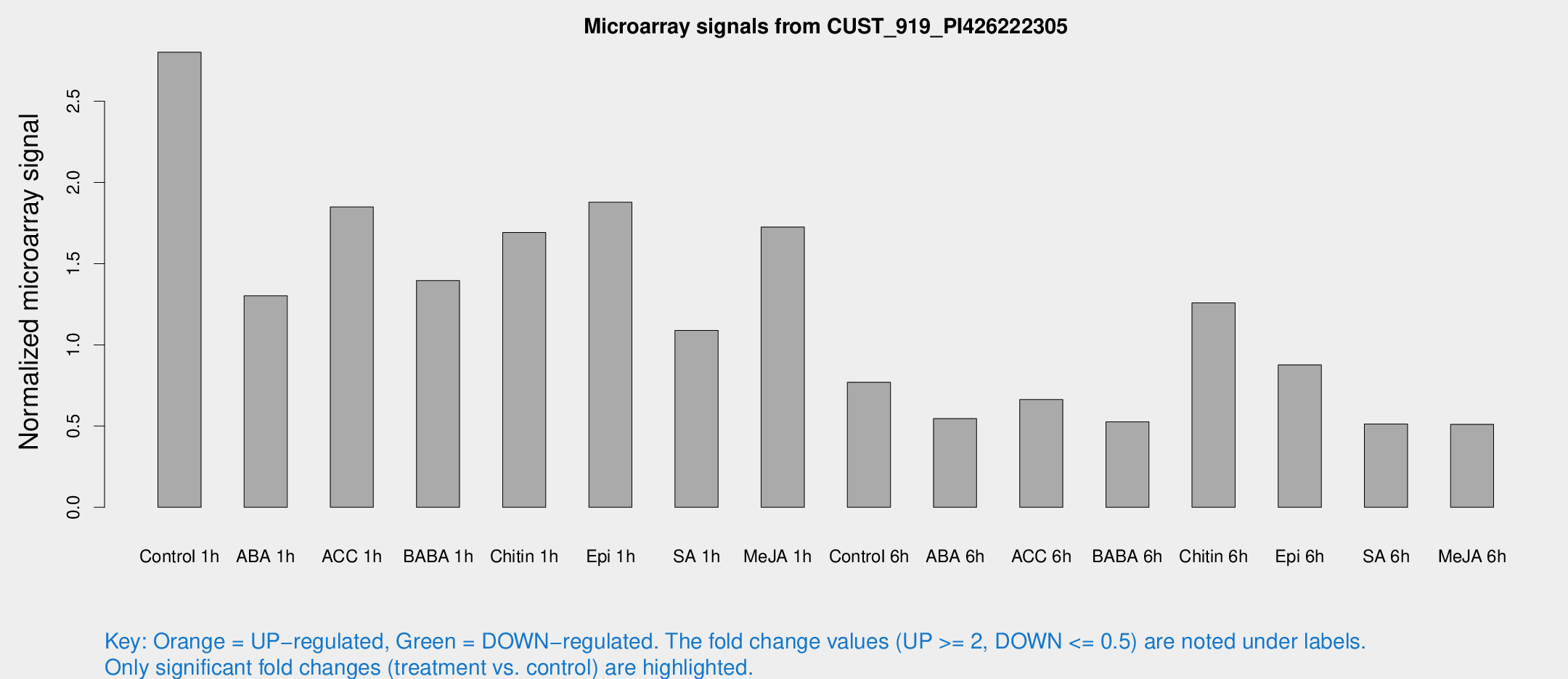
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 43.6364 | 6.70593 | 2.80138 | 0.356649 |
| ABA 1h | 24.2089 | 11.8396 | 1.30159 | 1.18835 |
| ACC 1h | 35.9524 | 13.7979 | 1.84894 | 0.945801 |
| BABA 1h | 20.9775 | 3.8477 | 1.39559 | 0.276787 |
| Chitin 1h | 25.4311 | 7.97488 | 1.69176 | 0.65012 |
| Epi 1h | 25.8684 | 5.31462 | 1.87802 | 0.410346 |
| SA 1h | 18.1141 | 4.75519 | 1.08933 | 0.365914 |
| Me-JA 1h | 21.8975 | 3.75479 | 1.72561 | 0.307904 |
| Control 6h | 12.8987 | 3.6277 | 0.769766 | 0.323252 |
| ABA 6h | 9.58133 | 3.67835 | 0.545803 | 0.244737 |
| ACC 6h | 12.7355 | 4.21498 | 0.663438 | 0.240343 |
| BABA 6h | 9.97781 | 3.98589 | 0.525953 | 0.252742 |
| Chitin 6h | 20.3263 | 4.08075 | 1.25824 | 0.252418 |
| Epi 6h | 15.176 | 4.2641 | 0.876858 | 0.243059 |
| SA 6h | 7.87604 | 3.80199 | 0.512498 | 0.260701 |
| Me-JA 6h | 8.18028 | 3.5532 | 0.510861 | 0.248796 |
Source Transcript PGSC0003DMT400025952 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g090540.2 | +1 | 2e-153 | 431 | 239/260 (92%) | genomic_reference:SL2.50ch02 gene_region:46644757-46646996 transcript_region:SL2.50ch02:46644757..46646996- go_terms:GO:0050327 functional_description:Short-chain dehydrogenase/reductase family protein (AHRD V1 **** D7LTG0_ARALY); contains Interpro domain(s) IPR002347 Glucose/ribitol dehydrogenase |
| TAIR PP10 | AT3G50560.1 | +1 | 1e-107 | 316 | 184/271 (68%) | NAD(P)-binding Rossmann-fold superfamily protein | chr3:18761247-18763835 REVERSE LENGTH=272 |
