Probe CUST_9183_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9183_PI426222305 | JHI_St_60k_v1 | DMT400006652 | GATACTGCAGTTATAACAAAAGAAGCACATTGAGACTGATTCTTGATTAGTCCATGTGTA |
All Microarray Probes Designed to Gene DMG400002590
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9170_PI426222305 | JHI_St_60k_v1 | DMT400006656 | GAATTACATTGGATCAGCCACTTGTTTATGATTGTTTTTCTGTCTTCAGTGTCAAGATTG |
| CUST_9183_PI426222305 | JHI_St_60k_v1 | DMT400006652 | GATACTGCAGTTATAACAAAAGAAGCACATTGAGACTGATTCTTGATTAGTCCATGTGTA |
| CUST_9382_PI426222305 | JHI_St_60k_v1 | DMT400006654 | GATACTGCAGTTATAACAAAAGAAGCACATTGAGACTGATTCTTGATTAGTCCATGTGTA |
Microarray Signals from CUST_9183_PI426222305
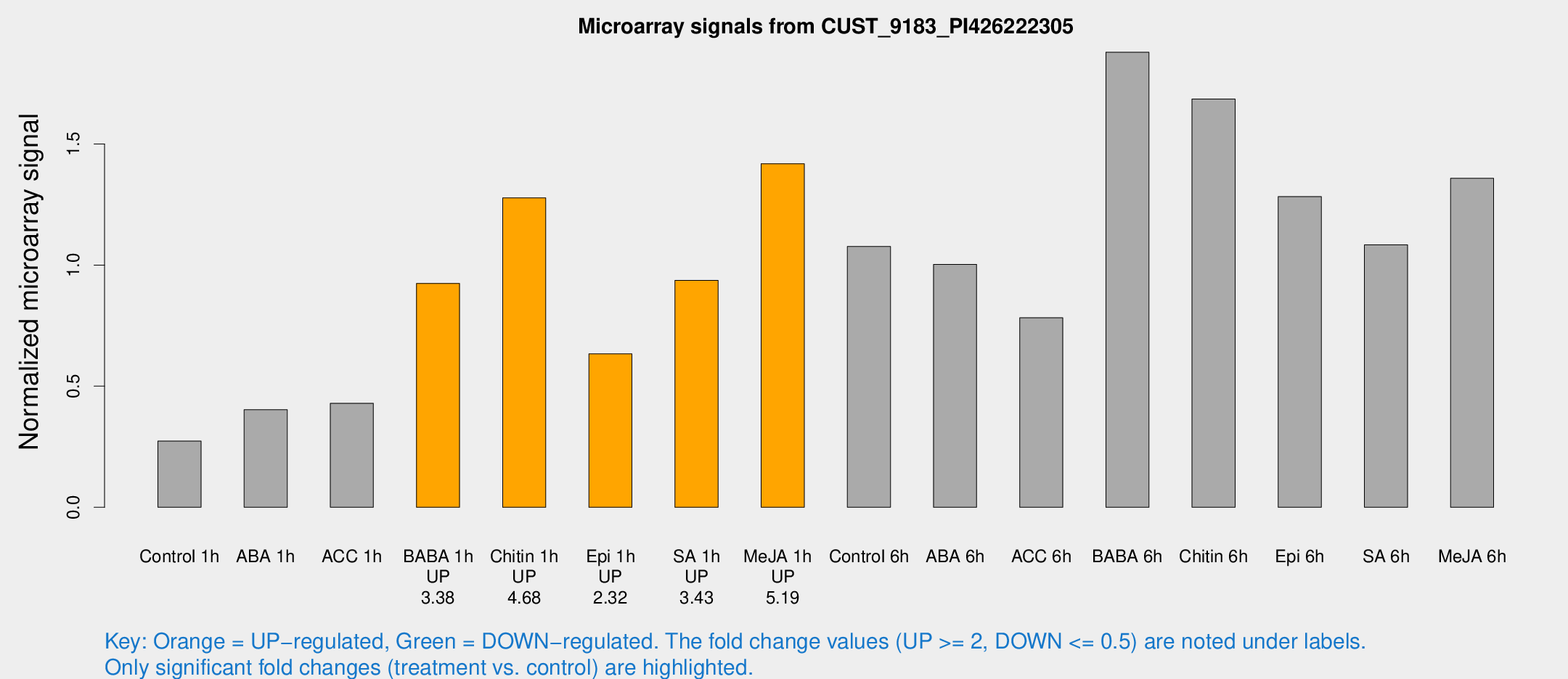
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1278.18 | 73.9408 | 0.273166 | 0.0189828 |
| ABA 1h | 1793.54 | 511.439 | 0.402944 | 0.0833813 |
| ACC 1h | 2091.57 | 241.622 | 0.429346 | 0.0248032 |
| BABA 1h | 4370.84 | 877.225 | 0.924396 | 0.11614 |
| Chitin 1h | 5407.13 | 450.442 | 1.27719 | 0.184677 |
| Epi 1h | 2626.72 | 415.056 | 0.633412 | 0.0837379 |
| SA 1h | 4504.49 | 260.484 | 0.93647 | 0.0540722 |
| Me-JA 1h | 5456.29 | 489.576 | 1.41856 | 0.0819062 |
| Control 6h | 5406.24 | 1262.77 | 1.07648 | 0.204512 |
| ABA 6h | 5065.75 | 664.986 | 1.00282 | 0.0769749 |
| ACC 6h | 4535.27 | 1327.76 | 0.782599 | 0.109337 |
| BABA 6h | 10184.1 | 1764.19 | 1.87864 | 0.297785 |
| Chitin 6h | 8496.49 | 986.358 | 1.6856 | 0.139073 |
| Epi 6h | 6944.27 | 1063.22 | 1.28234 | 0.316275 |
| SA 6h | 5042.32 | 392.639 | 1.08337 | 0.0625545 |
| Me-JA 6h | 7695.35 | 2686.02 | 1.35816 | 0.65252 |
Source Transcript PGSC0003DMT400006652 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G13750.1 | +2 | 2e-119 | 352 | 154/218 (71%) | beta galactosidase 1 | chr3:4511192-4515756 FORWARD LENGTH=847 |
