Probe CUST_9167_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9167_PI426222305 | JHI_St_60k_v1 | DMT400058932 | GGTGGACTAGTGAGTCTTCTCTGCATATATGTAATATCAAGATGTATGTTTGTATCTACA |
All Microarray Probes Designed to Gene DMG400022894
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9333_PI426222305 | JHI_St_60k_v1 | DMT400058934 | GGTGGACTAGTGAGTCTTCTCTGCATATATGTAATATCAAGATGTATGTTTGTATCTACA |
| CUST_9393_PI426222305 | JHI_St_60k_v1 | DMT400058933 | GGTGGACTAGTGAGTCTTCTCTGCATATATGTAATATCAAGATGTATGTTTGTATCTACA |
| CUST_9167_PI426222305 | JHI_St_60k_v1 | DMT400058932 | GGTGGACTAGTGAGTCTTCTCTGCATATATGTAATATCAAGATGTATGTTTGTATCTACA |
Microarray Signals from CUST_9167_PI426222305
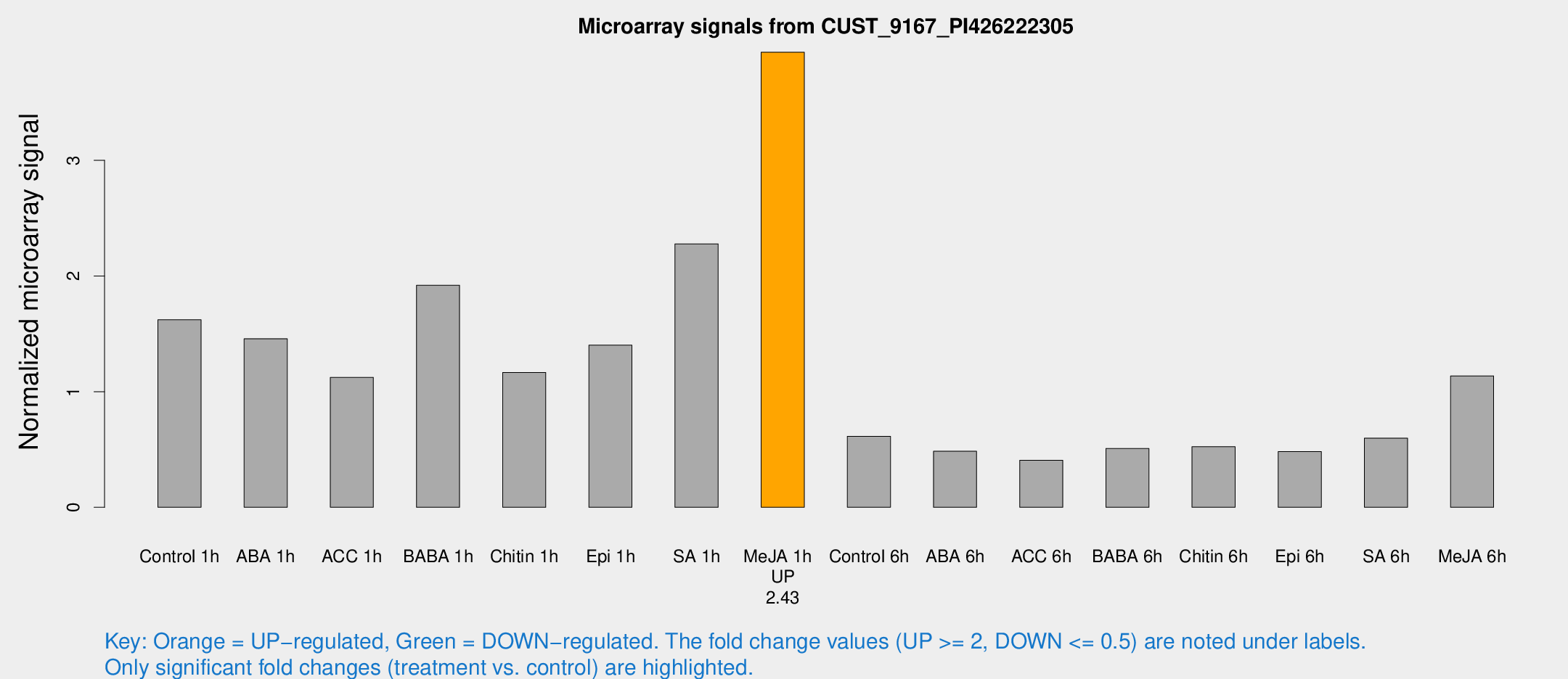
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 22967.3 | 3882.9 | 1.62124 | 0.258669 |
| ABA 1h | 19146.2 | 5350.43 | 1.45744 | 0.310553 |
| ACC 1h | 20987.3 | 8812.58 | 1.12283 | 0.720564 |
| BABA 1h | 26189.5 | 4132.84 | 1.92008 | 0.13219 |
| Chitin 1h | 14771.3 | 2270 | 1.16587 | 0.167425 |
| Epi 1h | 17406 | 3310.22 | 1.40177 | 0.291893 |
| SA 1h | 33336 | 6398.25 | 2.27789 | 0.403211 |
| Me-JA 1h | 44518.6 | 4058.84 | 3.93455 | 0.227162 |
| Control 6h | 8987.11 | 2025.69 | 0.614291 | 0.0990195 |
| ABA 6h | 7162.18 | 739.282 | 0.484348 | 0.0302915 |
| ACC 6h | 6539.54 | 937.217 | 0.406689 | 0.0234815 |
| BABA 6h | 7892.21 | 733.001 | 0.508512 | 0.0416833 |
| Chitin 6h | 7680.97 | 444.289 | 0.523793 | 0.0356864 |
| Epi 6h | 7621.2 | 1117.83 | 0.48182 | 0.0763493 |
| SA 6h | 8211.77 | 861.597 | 0.597321 | 0.0448792 |
| Me-JA 6h | 15711.4 | 1519.15 | 1.13599 | 0.065587 |
Source Transcript PGSC0003DMT400058932 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G17420.1 | +2 | 0.0 | 733 | 343/438 (78%) | lipoxygenase 3 | chr1:5977512-5981384 FORWARD LENGTH=919 |
