Probe CUST_9012_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9012_PI426222305 | JHI_St_60k_v1 | DMT400033370 | GTTGTTGAAACATTGTTTCCGGGACCTTCTGATGAGCATAGTACTATTTCTACTAGACTG |
All Microarray Probes Designed to Gene DMG400012819
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8936_PI426222305 | JHI_St_60k_v1 | DMT400033371 | TCTGTTATTCCTTTCCTATCAGCAGGTACCTCCATCTTTTGCCATCAAGTCATCAGATGA |
| CUST_9012_PI426222305 | JHI_St_60k_v1 | DMT400033370 | GTTGTTGAAACATTGTTTCCGGGACCTTCTGATGAGCATAGTACTATTTCTACTAGACTG |
Microarray Signals from CUST_9012_PI426222305
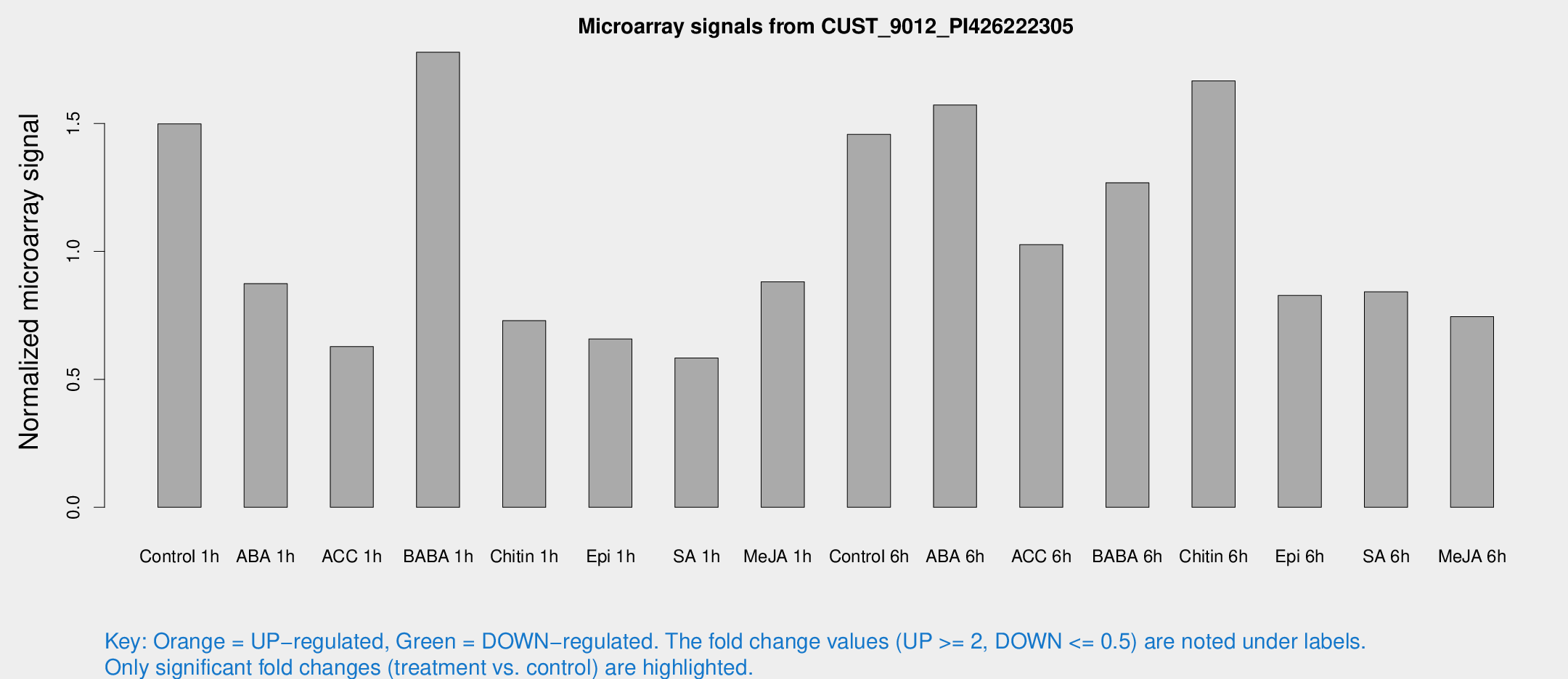
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 24.8014 | 3.76123 | 1.4984 | 0.259505 |
| ABA 1h | 14.3992 | 4.36238 | 0.873832 | 0.32209 |
| ACC 1h | 11.7157 | 4.50308 | 0.628131 | 0.292367 |
| BABA 1h | 37.3264 | 18.9335 | 1.7784 | 0.970207 |
| Chitin 1h | 12.0142 | 4.23786 | 0.729284 | 0.322707 |
| Epi 1h | 9.41315 | 3.43559 | 0.657986 | 0.243278 |
| SA 1h | 10.014 | 3.54752 | 0.583526 | 0.216681 |
| Me-JA 1h | 11.9727 | 3.67169 | 0.881506 | 0.277887 |
| Control 6h | 33.7292 | 14.4434 | 1.45708 | 1.13222 |
| ABA 6h | 29.8108 | 7.72638 | 1.57221 | 0.427691 |
| ACC 6h | 19.6126 | 4.59336 | 1.02664 | 0.246249 |
| BABA 6h | 24.8936 | 5.76218 | 1.26825 | 0.314955 |
| Chitin 6h | 29.5668 | 4.28321 | 1.66665 | 0.277998 |
| Epi 6h | 16.6904 | 4.61912 | 0.827832 | 0.32309 |
| SA 6h | 16.8343 | 8.04034 | 0.842233 | 0.610821 |
| Me-JA 6h | 14.117 | 5.0745 | 0.745056 | 0.270137 |
Source Transcript PGSC0003DMT400033370 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G28530.1 | +1 | 2e-163 | 493 | 301/616 (49%) | unknown protein; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast; EXPRESSED IN: 19 plant structures; EXPRESSED DURING: 10 growth stages; Has 20 Blast hits to 20 proteins in 6 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 20; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr1:10032127-10035496 REVERSE LENGTH=614 |
