Probe CUST_8936_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8936_PI426222305 | JHI_St_60k_v1 | DMT400033371 | TCTGTTATTCCTTTCCTATCAGCAGGTACCTCCATCTTTTGCCATCAAGTCATCAGATGA |
All Microarray Probes Designed to Gene DMG400012819
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8936_PI426222305 | JHI_St_60k_v1 | DMT400033371 | TCTGTTATTCCTTTCCTATCAGCAGGTACCTCCATCTTTTGCCATCAAGTCATCAGATGA |
| CUST_9012_PI426222305 | JHI_St_60k_v1 | DMT400033370 | GTTGTTGAAACATTGTTTCCGGGACCTTCTGATGAGCATAGTACTATTTCTACTAGACTG |
Microarray Signals from CUST_8936_PI426222305
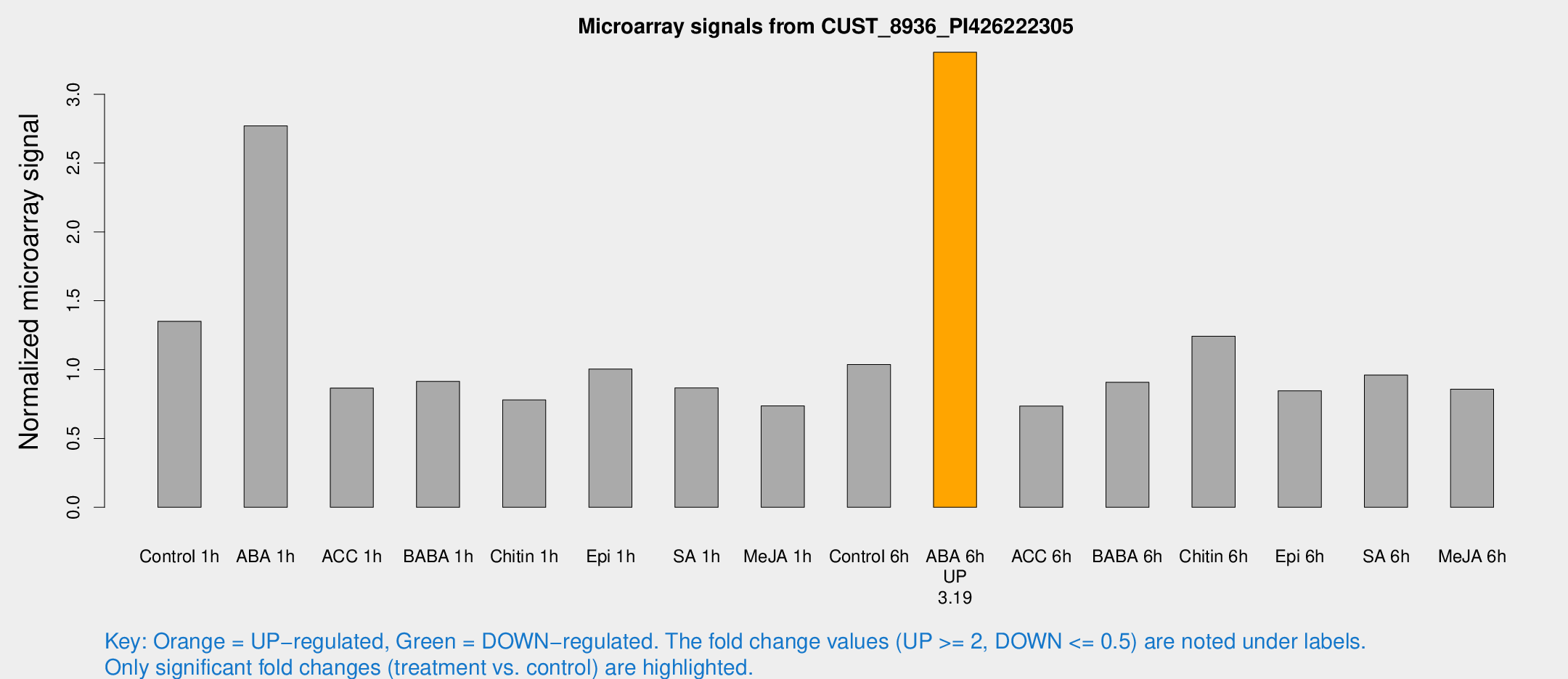
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 144.822 | 25.1583 | 1.35109 | 0.172748 |
| ABA 1h | 265.391 | 58.1335 | 2.76974 | 0.599655 |
| ACC 1h | 98.6434 | 26.7353 | 0.866005 | 0.195608 |
| BABA 1h | 98.7847 | 25.8866 | 0.915134 | 0.181348 |
| Chitin 1h | 76.7737 | 17.8145 | 0.780212 | 0.122713 |
| Epi 1h | 90.8691 | 8.26812 | 1.00431 | 0.0754988 |
| SA 1h | 93.645 | 11.4012 | 0.866993 | 0.0611397 |
| Me-JA 1h | 63.8673 | 10.3679 | 0.736598 | 0.0620905 |
| Control 6h | 112.287 | 21.1974 | 1.03709 | 0.129678 |
| ABA 6h | 373.795 | 61.4616 | 3.30508 | 0.350257 |
| ACC 6h | 94.8656 | 24.7139 | 0.73496 | 0.214659 |
| BABA 6h | 107.011 | 13.1663 | 0.90787 | 0.0906521 |
| Chitin 6h | 139.294 | 17.7538 | 1.24302 | 0.131783 |
| Epi 6h | 101.13 | 14.767 | 0.846236 | 0.0638212 |
| SA 6h | 102.741 | 21.9257 | 0.960509 | 0.199413 |
| Me-JA 6h | 92.7096 | 19.6301 | 0.858383 | 0.112848 |
Source Transcript PGSC0003DMT400033371 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G28530.1 | +2 | 7e-55 | 199 | 112/241 (46%) | unknown protein; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast; EXPRESSED IN: 19 plant structures; EXPRESSED DURING: 10 growth stages; Has 20 Blast hits to 20 proteins in 6 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 20; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr1:10032127-10035496 REVERSE LENGTH=614 |
