Probe CUST_7691_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7691_PI426222305 | JHI_St_60k_v1 | DMT400009553 | CAACTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTA |
All Microarray Probes Designed to Gene DMG400003726
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7471_PI426222305 | JHI_St_60k_v1 | DMT400009551 | CTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTAGAT |
| CUST_7691_PI426222305 | JHI_St_60k_v1 | DMT400009553 | CAACTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTA |
| CUST_7473_PI426222305 | JHI_St_60k_v1 | DMT400009552 | CTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTAGAT |
Microarray Signals from CUST_7691_PI426222305
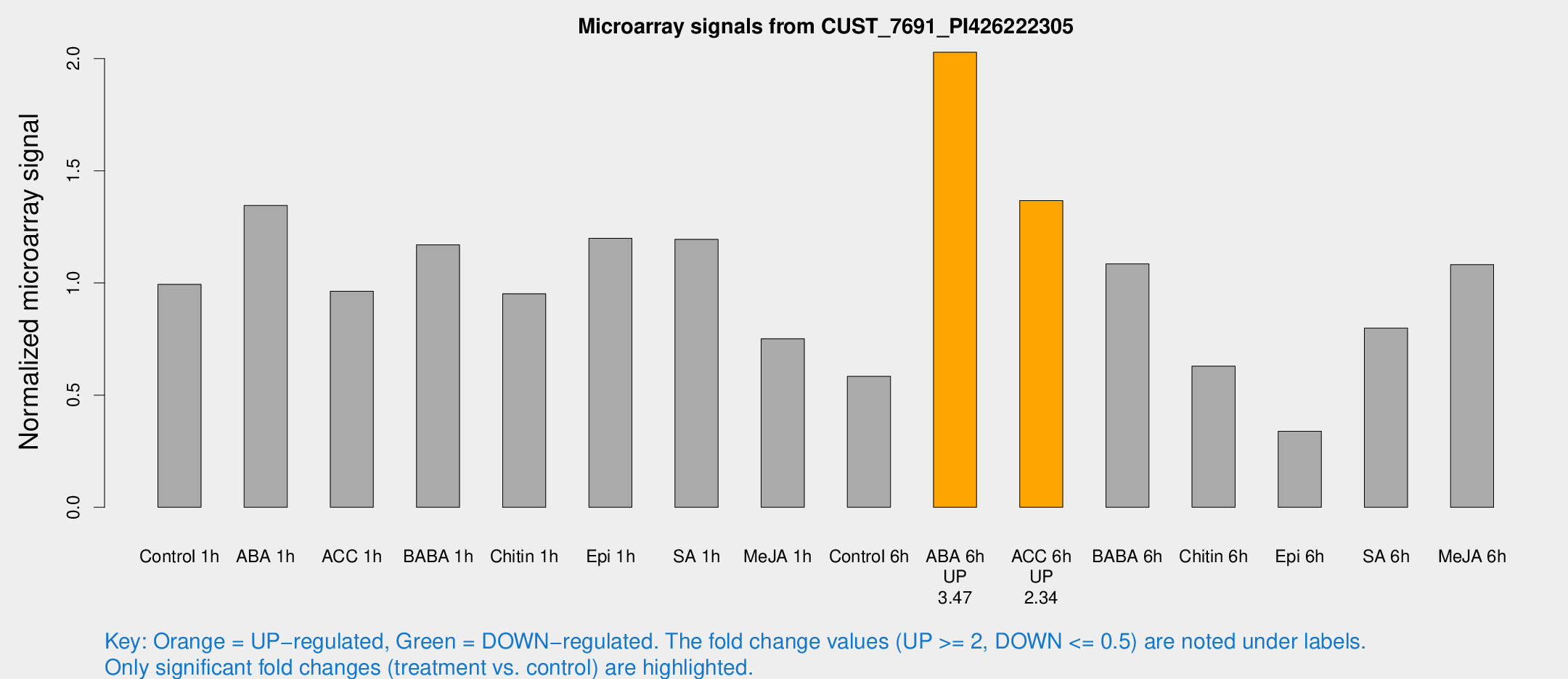
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3939.24 | 227.707 | 0.993398 | 0.0573585 |
| ABA 1h | 4762.68 | 473.777 | 1.34547 | 0.0776849 |
| ACC 1h | 4364.85 | 1217.9 | 0.963074 | 0.28047 |
| BABA 1h | 4772.04 | 1138.38 | 1.1698 | 0.199324 |
| Chitin 1h | 3444.93 | 446.552 | 0.952086 | 0.0675874 |
| Epi 1h | 4119.81 | 238.462 | 1.1987 | 0.0692126 |
| SA 1h | 5144.78 | 1102.16 | 1.19438 | 0.241423 |
| Me-JA 1h | 2442.22 | 171.467 | 0.751308 | 0.0433868 |
| Control 6h | 2396.16 | 403.439 | 0.584151 | 0.0530073 |
| ABA 6h | 8820.65 | 1582.98 | 2.02847 | 0.252397 |
| ACC 6h | 6325.69 | 849.191 | 1.3674 | 0.298741 |
| BABA 6h | 4882.56 | 599.965 | 1.08474 | 0.124754 |
| Chitin 6h | 2704.89 | 359.738 | 0.629301 | 0.110981 |
| Epi 6h | 1594.73 | 329.579 | 0.339312 | 0.0559489 |
| SA 6h | 3169.85 | 345.128 | 0.798711 | 0.0461216 |
| Me-JA 6h | 4316.12 | 468.435 | 1.08186 | 0.094647 |
Source Transcript PGSC0003DMT400009553 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G28305.1 | +3 | 6e-18 | 85 | 42/71 (59%) | Putative lysine decarboxylase family protein | chr2:12081186-12084307 FORWARD LENGTH=213 |
