Probe CUST_7612_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7612_PI426222305 | JHI_St_60k_v1 | DMT400025845 | TCCAAATTCCTTCAAAGTGCTGTCTCCTGCGCGAACATGTGTGGCTTATTACAGAGTTGA |
All Microarray Probes Designed to Gene DMG400009981
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7612_PI426222305 | JHI_St_60k_v1 | DMT400025845 | TCCAAATTCCTTCAAAGTGCTGTCTCCTGCGCGAACATGTGTGGCTTATTACAGAGTTGA |
| CUST_7418_PI426222305 | JHI_St_60k_v1 | DMT400025846 | ATTCCTTCAAAGTGCTGTCTCCTGCGCGAACATGTGTGGCTTATTACAGAGTTGATGAAC |
| CUST_7544_PI426222305 | JHI_St_60k_v1 | DMT400025848 | GTGCTTTGCCTTTGGAGCCAAGTCTCTATATGTAATAAGAAAACTAAGAACAATCACATA |
| CUST_7752_PI426222305 | JHI_St_60k_v1 | DMT400025844 | TTGGTGCTTTGCCTTTGGAGCCAAGTCTCTATATGTAATAAGAAAACTAAGAACAATCAC |
Microarray Signals from CUST_7612_PI426222305
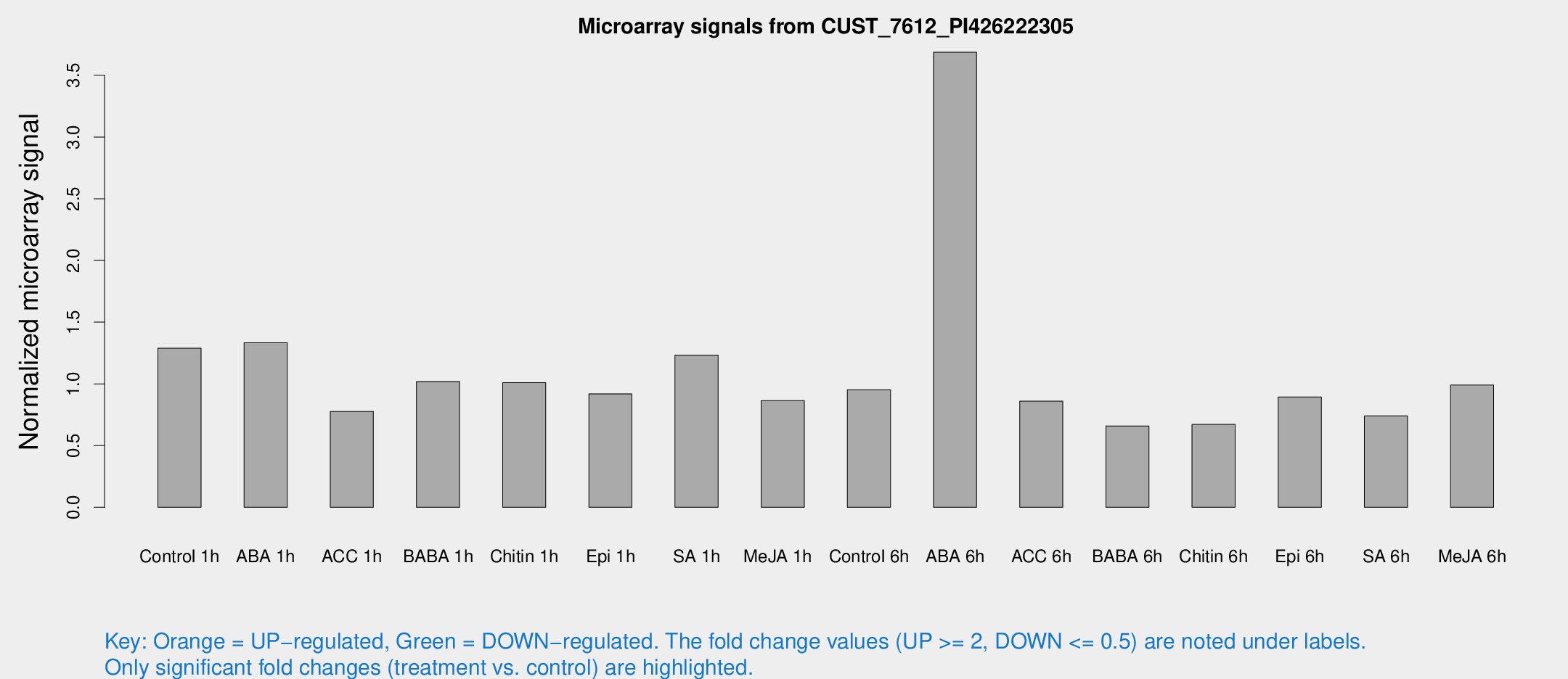
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 872.496 | 57.8685 | 1.28859 | 0.0897411 |
| ABA 1h | 797.13 | 46.2375 | 1.33391 | 0.132981 |
| ACC 1h | 638.679 | 212.576 | 0.776734 | 0.329501 |
| BABA 1h | 720.145 | 185.409 | 1.01919 | 0.211627 |
| Chitin 1h | 652.36 | 170.905 | 1.00987 | 0.20344 |
| Epi 1h | 557.53 | 104.426 | 0.919251 | 0.183197 |
| SA 1h | 898.974 | 185.827 | 1.23287 | 0.240765 |
| Me-JA 1h | 504.483 | 118.282 | 0.864823 | 0.188132 |
| Control 6h | 701.195 | 206.655 | 0.952467 | 0.231611 |
| ABA 6h | 2800.71 | 636.346 | 3.6871 | 0.837093 |
| ACC 6h | 665.134 | 38.6454 | 0.860004 | 0.112865 |
| BABA 6h | 505.144 | 64.3883 | 0.659037 | 0.0897388 |
| Chitin 6h | 488.624 | 54.1901 | 0.672261 | 0.0668927 |
| Epi 6h | 779.141 | 242.035 | 0.893613 | 0.318968 |
| SA 6h | 519.683 | 112.66 | 0.74102 | 0.0946341 |
| Me-JA 6h | 674.144 | 81.9224 | 0.990346 | 0.0574148 |
Source Transcript PGSC0003DMT400025845 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G36390.1 | +3 | 0.0 | 819 | 401/695 (58%) | starch branching enzyme 2.1 | chr2:15264283-15269940 FORWARD LENGTH=858 |
