Probe CUST_7473_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7473_PI426222305 | JHI_St_60k_v1 | DMT400009552 | CTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTAGAT |
All Microarray Probes Designed to Gene DMG400003726
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7471_PI426222305 | JHI_St_60k_v1 | DMT400009551 | CTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTAGAT |
| CUST_7691_PI426222305 | JHI_St_60k_v1 | DMT400009553 | CAACTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTA |
| CUST_7473_PI426222305 | JHI_St_60k_v1 | DMT400009552 | CTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTAGAT |
Microarray Signals from CUST_7473_PI426222305
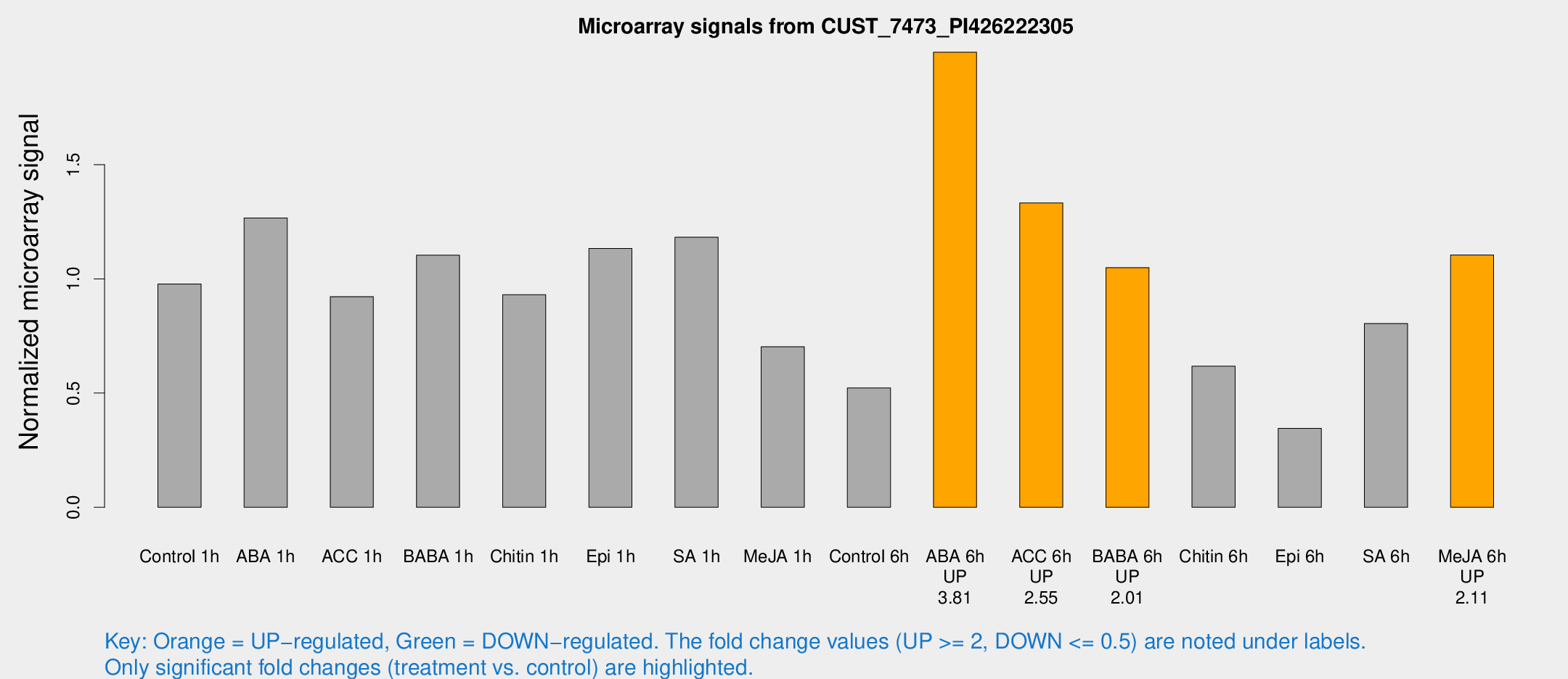
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4050.81 | 234.266 | 0.977862 | 0.0564622 |
| ABA 1h | 4701.48 | 552.679 | 1.26618 | 0.0767365 |
| ACC 1h | 4425.21 | 1295.58 | 0.921984 | 0.295998 |
| BABA 1h | 4806.7 | 1287.71 | 1.10352 | 0.239972 |
| Chitin 1h | 3540.77 | 553.235 | 0.930711 | 0.0781815 |
| Epi 1h | 4063.36 | 235.021 | 1.13307 | 0.0654238 |
| SA 1h | 5258.99 | 1050.23 | 1.18217 | 0.195243 |
| Me-JA 1h | 2387.2 | 178.696 | 0.702799 | 0.040588 |
| Control 6h | 2232.31 | 362.083 | 0.522432 | 0.0438179 |
| ABA 6h | 9016.6 | 1549.57 | 1.99211 | 0.228059 |
| ACC 6h | 6409.14 | 751.658 | 1.33279 | 0.280619 |
| BABA 6h | 4932.24 | 607.81 | 1.04924 | 0.122773 |
| Chitin 6h | 2794.47 | 444.326 | 0.617685 | 0.126946 |
| Epi 6h | 1692.26 | 349.264 | 0.345369 | 0.0545148 |
| SA 6h | 3303.73 | 202.385 | 0.804377 | 0.0477752 |
| Me-JA 6h | 4572.94 | 328.552 | 1.10478 | 0.0637896 |
Source Transcript PGSC0003DMT400009552 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G06300.1 | +2 | 4e-88 | 274 | 155/179 (87%) | Putative lysine decarboxylase family protein | chr5:1922042-1925278 REVERSE LENGTH=217 |
