Probe CUST_7471_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7471_PI426222305 | JHI_St_60k_v1 | DMT400009551 | CTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTAGAT |
All Microarray Probes Designed to Gene DMG400003726
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7471_PI426222305 | JHI_St_60k_v1 | DMT400009551 | CTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTAGAT |
| CUST_7691_PI426222305 | JHI_St_60k_v1 | DMT400009553 | CAACTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTA |
| CUST_7473_PI426222305 | JHI_St_60k_v1 | DMT400009552 | CTTGGCTACACAACAACAAAATTGGAAATTGCTCGTTGAATATTCAAATGATGGGTAGAT |
Microarray Signals from CUST_7471_PI426222305
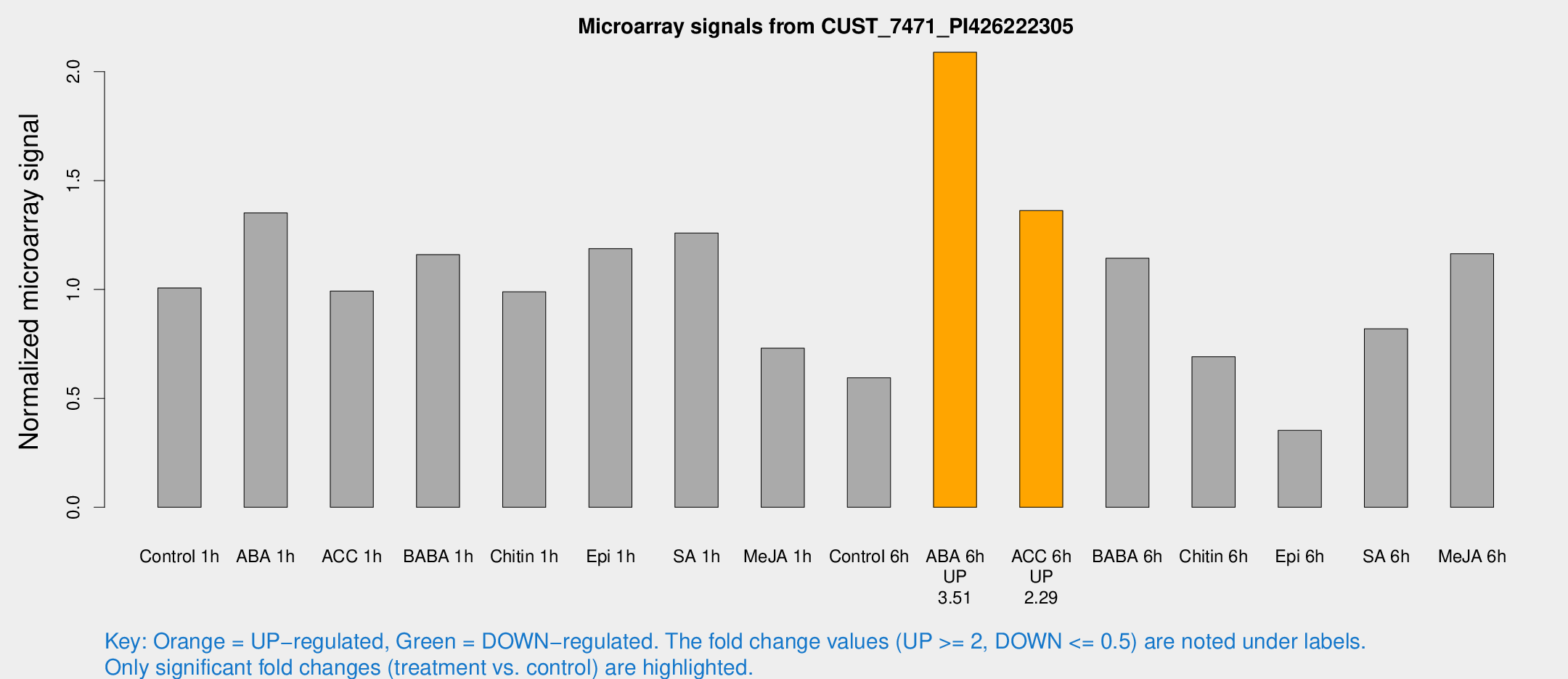
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3935.65 | 228.639 | 1.0072 | 0.0581565 |
| ABA 1h | 4723.84 | 547.968 | 1.35172 | 0.0780469 |
| ACC 1h | 4445.24 | 1268.46 | 0.992746 | 0.299706 |
| BABA 1h | 4674.53 | 1131.79 | 1.15971 | 0.209062 |
| Chitin 1h | 3532.7 | 511.137 | 0.98927 | 0.0805948 |
| Epi 1h | 4017.17 | 233.148 | 1.18762 | 0.0685735 |
| SA 1h | 5219.74 | 918.419 | 1.25899 | 0.175117 |
| Me-JA 1h | 2334.43 | 170.942 | 0.730167 | 0.0421683 |
| Control 6h | 2421.32 | 480.935 | 0.594346 | 0.0713567 |
| ABA 6h | 8939.89 | 1661.9 | 2.08912 | 0.262774 |
| ACC 6h | 6187.07 | 799.923 | 1.36241 | 0.299611 |
| BABA 6h | 5117.96 | 825.777 | 1.14314 | 0.182709 |
| Chitin 6h | 2922.67 | 397.601 | 0.691572 | 0.121266 |
| Epi 6h | 1614.33 | 302.352 | 0.353017 | 0.0478914 |
| SA 6h | 3175.62 | 237.995 | 0.819463 | 0.0473207 |
| Me-JA 6h | 4569.93 | 518.68 | 1.16391 | 0.0987623 |
Source Transcript PGSC0003DMT400009551 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G28305.1 | +2 | 1e-99 | 300 | 167/211 (79%) | Putative lysine decarboxylase family protein | chr2:12081186-12084307 FORWARD LENGTH=213 |
