Probe CUST_7418_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7418_PI426222305 | JHI_St_60k_v1 | DMT400025846 | ATTCCTTCAAAGTGCTGTCTCCTGCGCGAACATGTGTGGCTTATTACAGAGTTGATGAAC |
All Microarray Probes Designed to Gene DMG400009981
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_7612_PI426222305 | JHI_St_60k_v1 | DMT400025845 | TCCAAATTCCTTCAAAGTGCTGTCTCCTGCGCGAACATGTGTGGCTTATTACAGAGTTGA |
| CUST_7418_PI426222305 | JHI_St_60k_v1 | DMT400025846 | ATTCCTTCAAAGTGCTGTCTCCTGCGCGAACATGTGTGGCTTATTACAGAGTTGATGAAC |
| CUST_7544_PI426222305 | JHI_St_60k_v1 | DMT400025848 | GTGCTTTGCCTTTGGAGCCAAGTCTCTATATGTAATAAGAAAACTAAGAACAATCACATA |
| CUST_7752_PI426222305 | JHI_St_60k_v1 | DMT400025844 | TTGGTGCTTTGCCTTTGGAGCCAAGTCTCTATATGTAATAAGAAAACTAAGAACAATCAC |
Microarray Signals from CUST_7418_PI426222305
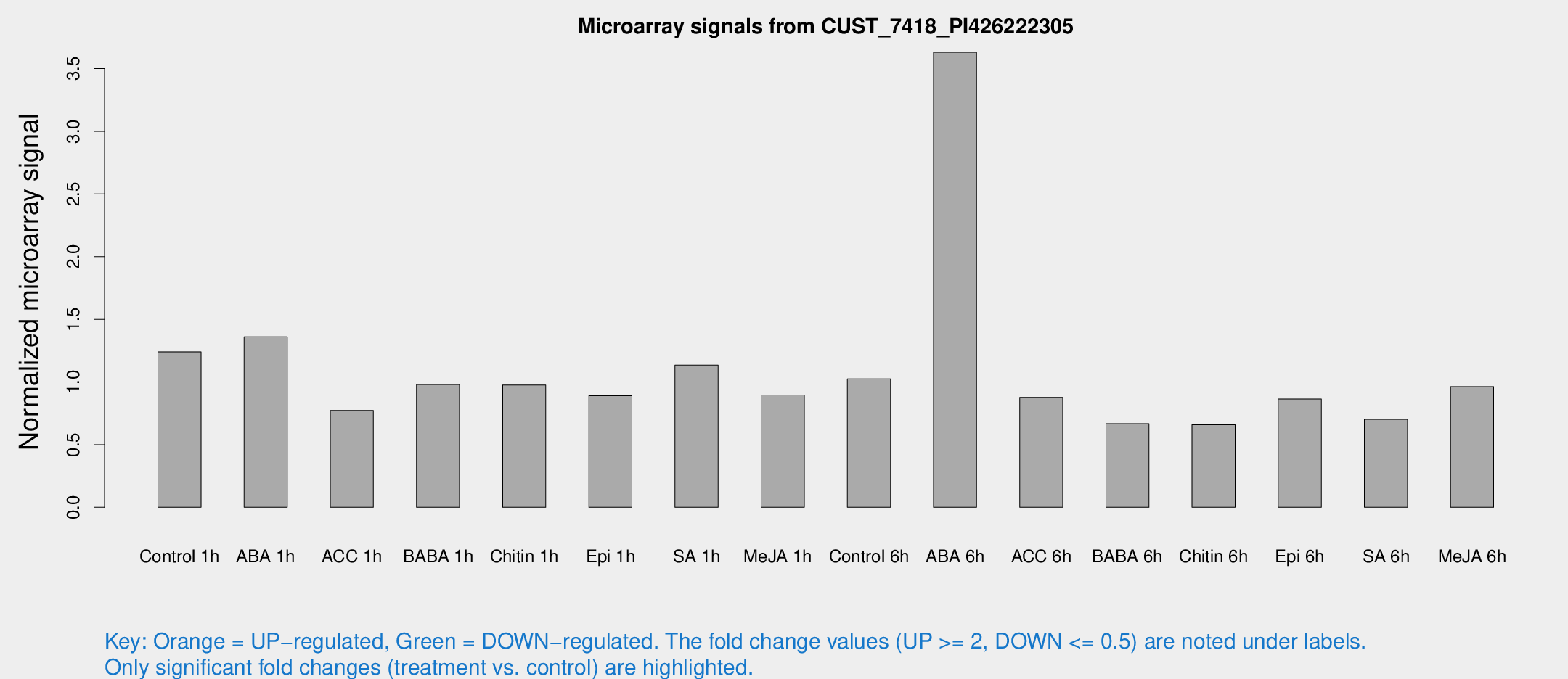
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1009.55 | 63.5686 | 1.24094 | 0.0794357 |
| ABA 1h | 976.469 | 56.5031 | 1.36098 | 0.136926 |
| ACC 1h | 752.895 | 242.494 | 0.772362 | 0.30537 |
| BABA 1h | 828.205 | 215.366 | 0.979984 | 0.193878 |
| Chitin 1h | 744.739 | 164.337 | 0.975871 | 0.168393 |
| Epi 1h | 659.503 | 155.378 | 0.890281 | 0.215623 |
| SA 1h | 995.451 | 213.259 | 1.13463 | 0.230532 |
| Me-JA 1h | 615.729 | 118.44 | 0.895623 | 0.153822 |
| Control 6h | 903.927 | 264.658 | 1.02479 | 0.246818 |
| ABA 6h | 3283.14 | 680.463 | 3.63056 | 0.733179 |
| ACC 6h | 815.712 | 47.2516 | 0.877409 | 0.129248 |
| BABA 6h | 613.958 | 76.2864 | 0.666737 | 0.0911066 |
| Chitin 6h | 579.144 | 78.554 | 0.659153 | 0.0834822 |
| Epi 6h | 879.074 | 247.854 | 0.864248 | 0.254702 |
| SA 6h | 614.189 | 171.249 | 0.702634 | 0.14188 |
| Me-JA 6h | 786.411 | 88.5207 | 0.962886 | 0.0557276 |
Source Transcript PGSC0003DMT400025846 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G36390.1 | +3 | 0.0 | 819 | 401/695 (58%) | starch branching enzyme 2.1 | chr2:15264283-15269940 FORWARD LENGTH=858 |
