Probe CUST_637_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_637_PI426222305 | JHI_St_60k_v1 | DMT400070567 | GAATGCATAGTTTGTCAATTTGGTTGCGAATGATTGAAATTTGTTCTATCGTGAGGTGTT |
All Microarray Probes Designed to Gene DMG400027428
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_637_PI426222305 | JHI_St_60k_v1 | DMT400070567 | GAATGCATAGTTTGTCAATTTGGTTGCGAATGATTGAAATTTGTTCTATCGTGAGGTGTT |
Microarray Signals from CUST_637_PI426222305
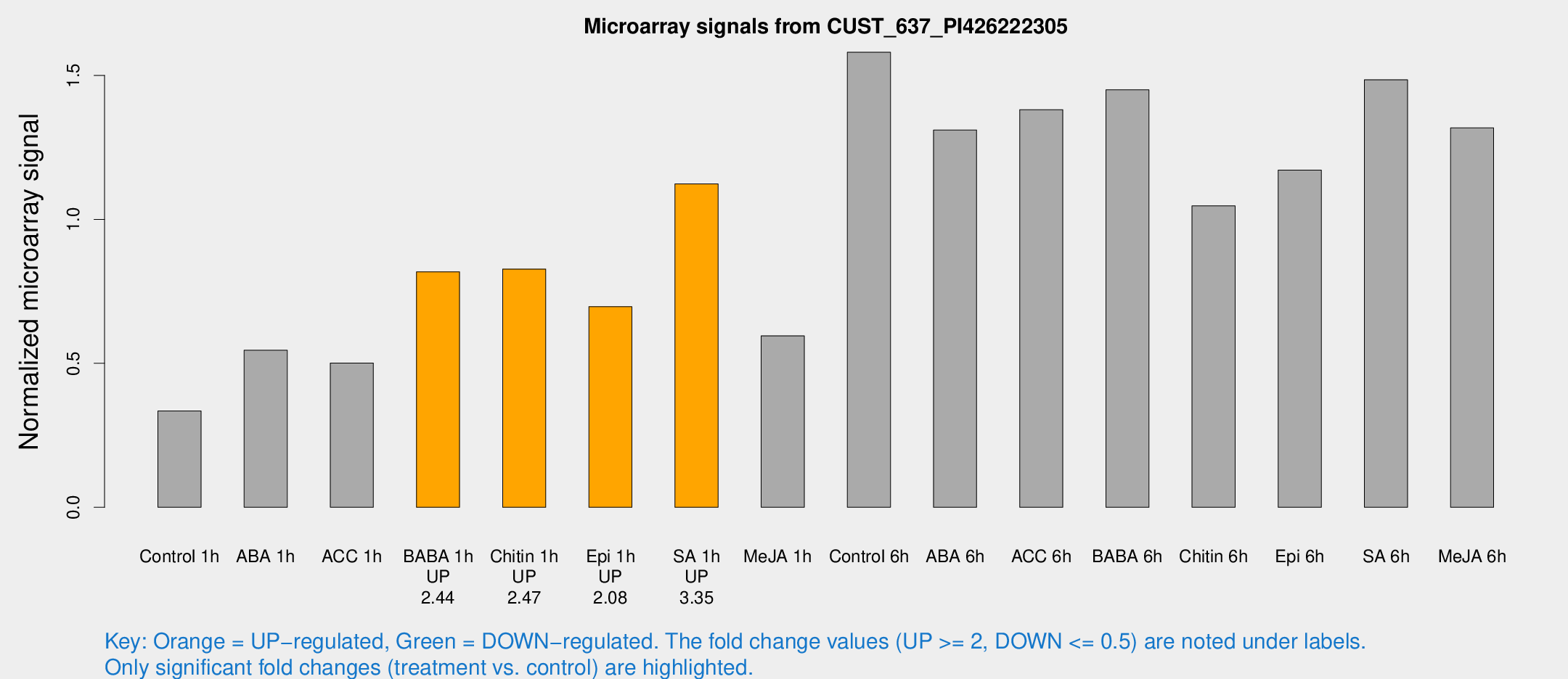
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 104.616 | 12.1843 | 0.334896 | 0.0223196 |
| ABA 1h | 150.18 | 14.6266 | 0.545395 | 0.0338789 |
| ACC 1h | 161.778 | 22.5279 | 0.500664 | 0.0436978 |
| BABA 1h | 252.275 | 46.6199 | 0.817798 | 0.0864633 |
| Chitin 1h | 235.436 | 36.8416 | 0.82734 | 0.110519 |
| Epi 1h | 192.255 | 36.3012 | 0.696912 | 0.107212 |
| SA 1h | 356.879 | 20.9664 | 1.1229 | 0.0996616 |
| Me-JA 1h | 150.458 | 9.43298 | 0.595528 | 0.0474653 |
| Control 6h | 519.66 | 117.445 | 1.58045 | 0.263107 |
| ABA 6h | 476.267 | 148.995 | 1.31027 | 0.373258 |
| ACC 6h | 497.184 | 66.9926 | 1.3809 | 0.0848023 |
| BABA 6h | 506.528 | 56.0442 | 1.44978 | 0.139874 |
| Chitin 6h | 344.639 | 20.3592 | 1.04704 | 0.061728 |
| Epi 6h | 410.952 | 38.0009 | 1.17104 | 0.189049 |
| SA 6h | 540.391 | 184.527 | 1.48475 | 0.556429 |
| Me-JA 6h | 420.275 | 75.1686 | 1.3176 | 0.174146 |
Source Transcript PGSC0003DMT400070567 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc00g170200.1 | +1 | 2e-80 | 246 | 118/128 (92%) | evidence_code:10F1H1E1IEG genomic_reference:SL2.50ch00 gene_region:18003903-18004286 transcript_region:SL2.50ch00:18003903..18004286+ functional_description:Hydrolase alpha/beta fold family protein (AHRD V1 ***- D7L528_ARALY) |
| TAIR PP10 | AT3G24420.1 | +1 | 3e-92 | 282 | 140/265 (53%) | alpha/beta-Hydrolases superfamily protein | chr3:8863111-8864883 REVERSE LENGTH=273 |
