Probe CUST_57_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_57_PI426222305 | JHI_St_60k_v1 | DMT400084231 | ATGCGCTGCACGAAGTCTTTTAGTTGATGATGAATAAACACAAGTGTTCATTGAGGATTA |
All Microarray Probes Designed to Gene DMG401033888
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_57_PI426222305 | JHI_St_60k_v1 | DMT400084231 | ATGCGCTGCACGAAGTCTTTTAGTTGATGATGAATAAACACAAGTGTTCATTGAGGATTA |
Microarray Signals from CUST_57_PI426222305
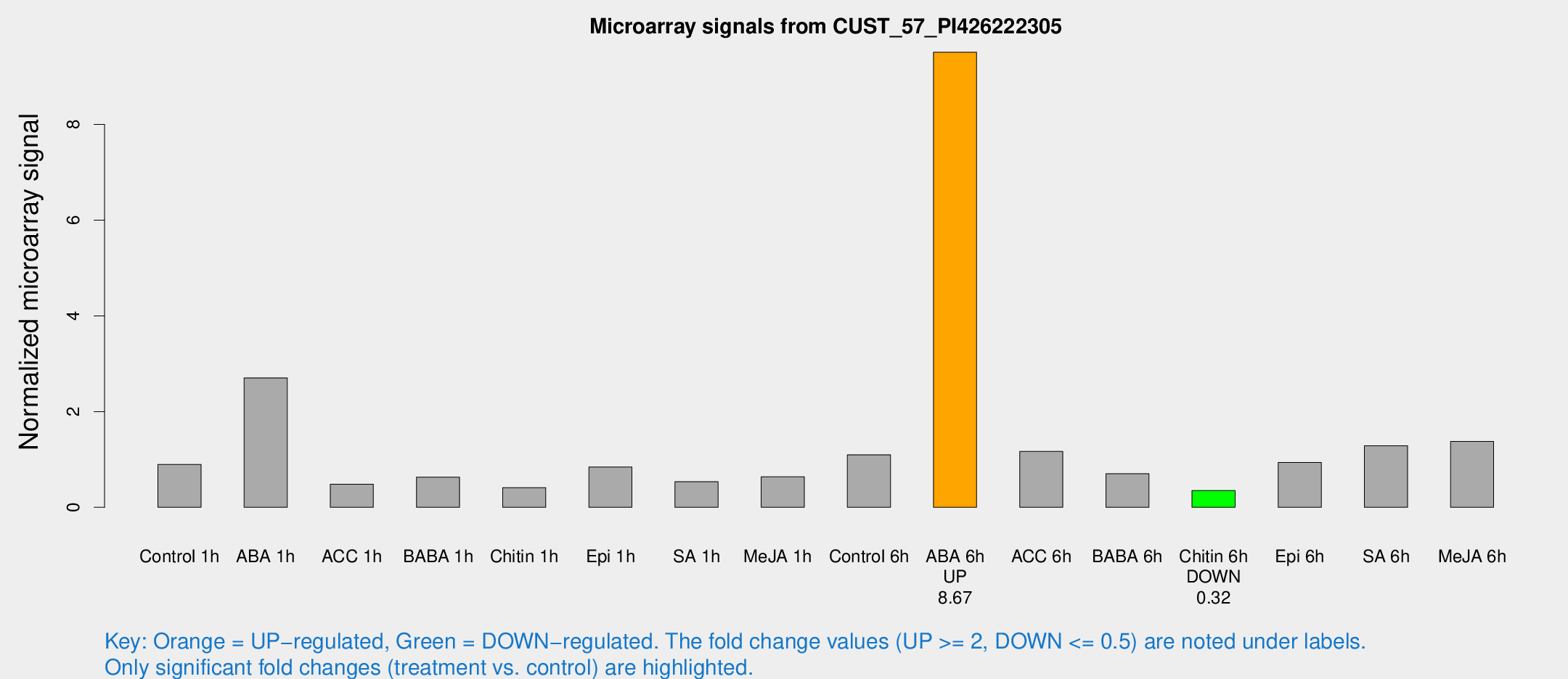
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 21.5858 | 3.65222 | 0.89483 | 0.180536 |
| ABA 1h | 59.808 | 14.4696 | 2.70393 | 0.697768 |
| ACC 1h | 13.3565 | 4.70487 | 0.483197 | 0.238444 |
| BABA 1h | 15.504 | 3.97986 | 0.62847 | 0.261281 |
| Chitin 1h | 9.07861 | 3.58311 | 0.41182 | 0.177018 |
| Epi 1h | 18.3337 | 4.26096 | 0.841333 | 0.245172 |
| SA 1h | 14.1842 | 4.21901 | 0.534177 | 0.161117 |
| Me-JA 1h | 13.6615 | 4.36227 | 0.636003 | 0.298952 |
| Control 6h | 26.9504 | 5.13034 | 1.09712 | 0.202065 |
| ABA 6h | 247.109 | 40.5668 | 9.50907 | 1.39768 |
| ACC 6h | 38.5208 | 13.3431 | 1.1685 | 0.614223 |
| BABA 6h | 21.4185 | 6.63661 | 0.700878 | 0.361699 |
| Chitin 6h | 9.14802 | 4.13853 | 0.350487 | 0.170077 |
| Epi 6h | 27.8533 | 7.76931 | 0.938728 | 0.271154 |
| SA 6h | 31.9123 | 7.02697 | 1.2844 | 0.203606 |
| Me-JA 6h | 33.1019 | 4.65619 | 1.37869 | 0.211012 |
Source Transcript PGSC0003DMT400084231 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc01g057000.2 | +1 | 7e-104 | 305 | 149/164 (91%) | genomic_reference:SL2.50ch01 gene_region:50798585-50800986 transcript_region:SL2.50ch01:50798585..50800986+ go_terms:GO:0006950 functional_description:Universal stress protein family protein (AHRD V1 ***- D7LAT3_ARALY); contains Interpro domain(s) IPR006016 UspA |
| TAIR PP10 | AT3G58450.1 | +1 | 8e-51 | 169 | 86/169 (51%) | Adenine nucleotide alpha hydrolases-like superfamily protein | chr3:21622032-21623057 FORWARD LENGTH=204 |
