Probe CUST_5415_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5415_PI426222305 | JHI_St_60k_v1 | DMT400009051 | CGGAACTTAACAATTTGGAGCTGATGAGCATGATATTCAACTGTAATGAAGTGTGTAAAT |
All Microarray Probes Designed to Gene DMG400003518
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5415_PI426222305 | JHI_St_60k_v1 | DMT400009051 | CGGAACTTAACAATTTGGAGCTGATGAGCATGATATTCAACTGTAATGAAGTGTGTAAAT |
Microarray Signals from CUST_5415_PI426222305
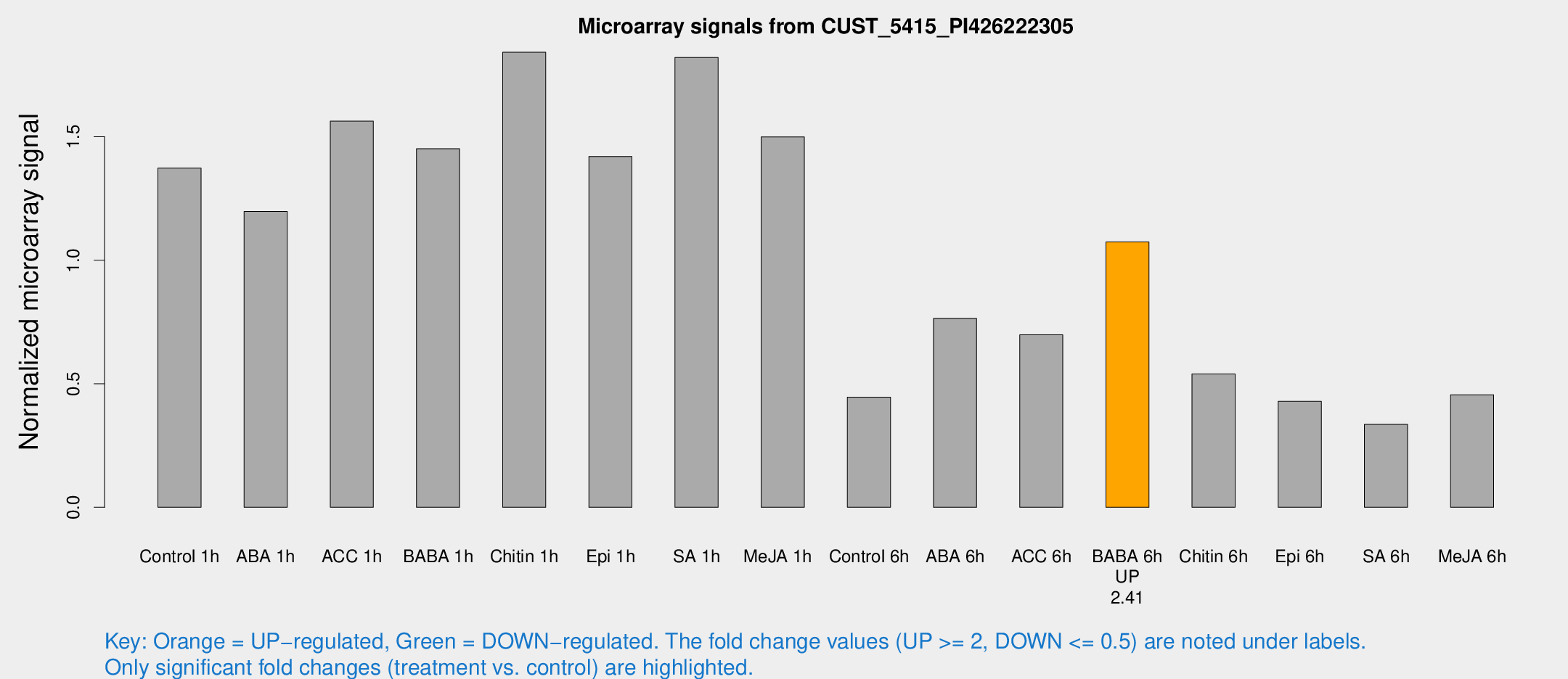
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1607.26 | 262.224 | 1.3733 | 0.213177 |
| ABA 1h | 1226.17 | 159.157 | 1.19802 | 0.217252 |
| ACC 1h | 1972.12 | 477.962 | 1.5635 | 0.34475 |
| BABA 1h | 1666.29 | 328.459 | 1.45156 | 0.172316 |
| Chitin 1h | 1887.14 | 109.08 | 1.84253 | 0.198424 |
| Epi 1h | 1415.72 | 154.413 | 1.42052 | 0.116678 |
| SA 1h | 2143.92 | 173.306 | 1.82142 | 0.118879 |
| Me-JA 1h | 1398.29 | 81.055 | 1.49991 | 0.0866649 |
| Control 6h | 527.567 | 102.373 | 0.445661 | 0.0514504 |
| ABA 6h | 949.033 | 147.822 | 0.764752 | 0.0812775 |
| ACC 6h | 943.272 | 181.089 | 0.698412 | 0.0404251 |
| BABA 6h | 1434.66 | 322.05 | 1.07425 | 0.195729 |
| Chitin 6h | 660.584 | 64.9722 | 0.539587 | 0.0608203 |
| Epi 6h | 555.673 | 54.3759 | 0.428973 | 0.0400841 |
| SA 6h | 384.776 | 51.317 | 0.335681 | 0.0213752 |
| Me-JA 6h | 539.172 | 114.244 | 0.45542 | 0.0581105 |
Source Transcript PGSC0003DMT400009051 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G36500.1 | +1 | 9e-13 | 63 | 44/112 (39%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: mitochondrion; EXPRESSED IN: 22 plant structures; EXPRESSED DURING: 13 growth stages; BEST Arabidopsis thaliana protein match is: unknown protein (TAIR:AT2G18210.1); Has 50 Blast hits to 50 proteins in 7 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 50; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr4:17226139-17226507 REVERSE LENGTH=122 |
